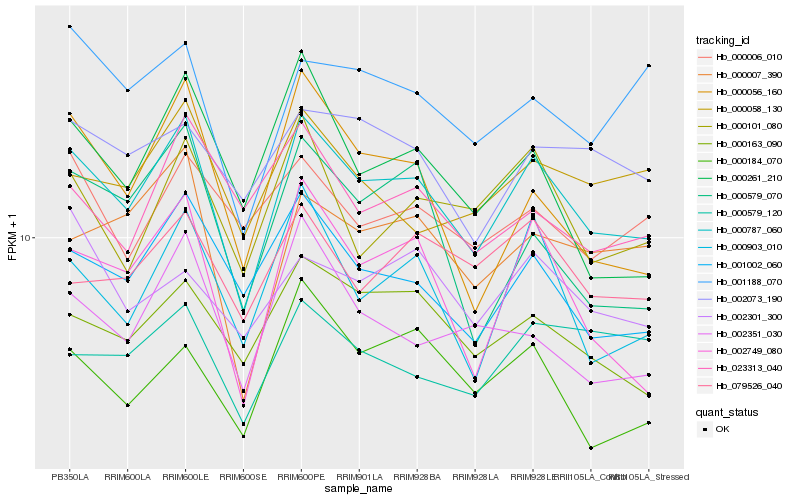
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000787_060 |
0.0 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_001002_060 |
0.079069091 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 3 |
Hb_000163_090 |
0.0813929469 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 4 |
Hb_000903_010 |
0.0819963595 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 5 |
Hb_002749_080 |
0.0825681371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000007_390 |
0.0852886501 |
- |
- |
hypothetical protein JCGZ_18250 [Jatropha curcas] |
| 7 |
Hb_000184_070 |
0.088301545 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000101_080 |
0.0903389112 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 9 |
Hb_000056_160 |
0.0915652087 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP23 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002351_030 |
0.0932180391 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_001188_070 |
0.0943368649 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 [Jatropha curcas] |
| 12 |
Hb_079526_040 |
0.0947195504 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 13 |
Hb_002073_190 |
0.0949768494 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000579_070 |
0.0958723014 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 15 |
Hb_000261_210 |
0.0962063363 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 16 |
Hb_000058_130 |
0.097221921 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 17 |
Hb_002301_300 |
0.0983899688 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 18 |
Hb_000579_120 |
0.0985082593 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 19 |
Hb_000006_010 |
0.0986045153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_023313_040 |
0.098858993 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |