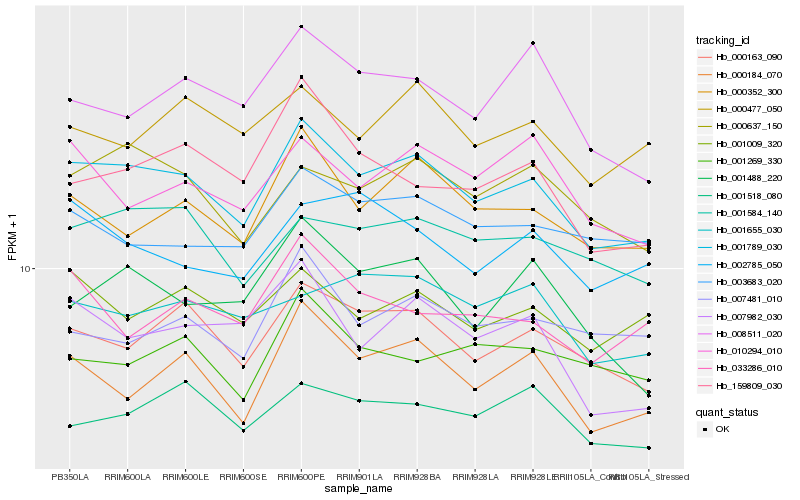
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001789_030 |
0.0 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 2 |
Hb_000163_090 |
0.0656751738 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 3 |
Hb_000352_300 |
0.0695786502 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 4 |
Hb_000184_070 |
0.0718142354 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000637_150 |
0.0761053117 |
- |
- |
Protein CASP, putative [Ricinus communis] |
| 6 |
Hb_001584_140 |
0.0773482219 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 7 |
Hb_001009_320 |
0.0801110526 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 8 |
Hb_007982_030 |
0.0834508228 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g10020 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003683_020 |
0.0843668528 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_010294_010 |
0.0850934811 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 11 |
Hb_000477_050 |
0.0853194925 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 12 |
Hb_001518_080 |
0.086070099 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 13 |
Hb_008511_020 |
0.0867856755 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 14 |
Hb_159809_030 |
0.0873332383 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001488_220 |
0.0876573265 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 16 |
Hb_007481_010 |
0.0884231358 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 17 |
Hb_002785_050 |
0.0893005999 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 18 |
Hb_033286_010 |
0.0896447111 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 19 |
Hb_001269_330 |
0.0897009465 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 20 |
Hb_001655_030 |
0.0903103304 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |