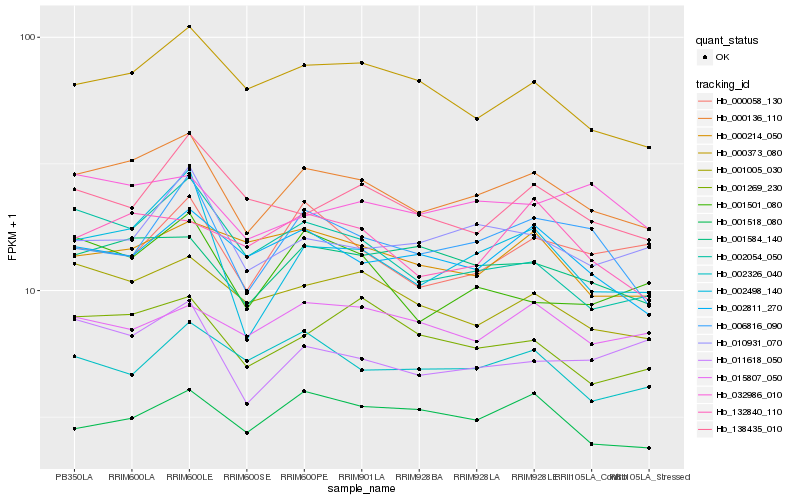
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000136_110 |
0.0 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 2 |
Hb_001584_140 |
0.0668173815 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 3 |
Hb_002498_140 |
0.0715797153 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 4 |
Hb_001005_030 |
0.0722978266 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000373_080 |
0.0765228718 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 6 |
Hb_002811_270 |
0.0769306364 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 7 |
Hb_001269_230 |
0.0770261908 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 8 |
Hb_010931_070 |
0.0800756284 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_132840_110 |
0.0827278968 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_001518_080 |
0.0828281882 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 11 |
Hb_006816_090 |
0.0828306501 |
- |
- |
PREDICTED: uncharacterized protein LOC105647425 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002054_050 |
0.0841373008 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 13 |
Hb_032986_010 |
0.0852499613 |
- |
- |
Nicotinate-nucleotide pyrophosphorylase [carboxylating], putative [Ricinus communis] |
| 14 |
Hb_011618_050 |
0.0854351692 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 15 |
Hb_000214_050 |
0.0856081249 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000058_130 |
0.0859688092 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 17 |
Hb_001501_080 |
0.0865270841 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002326_040 |
0.0872275527 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 19 |
Hb_138435_010 |
0.0874346157 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 20 |
Hb_015807_050 |
0.0881825242 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |