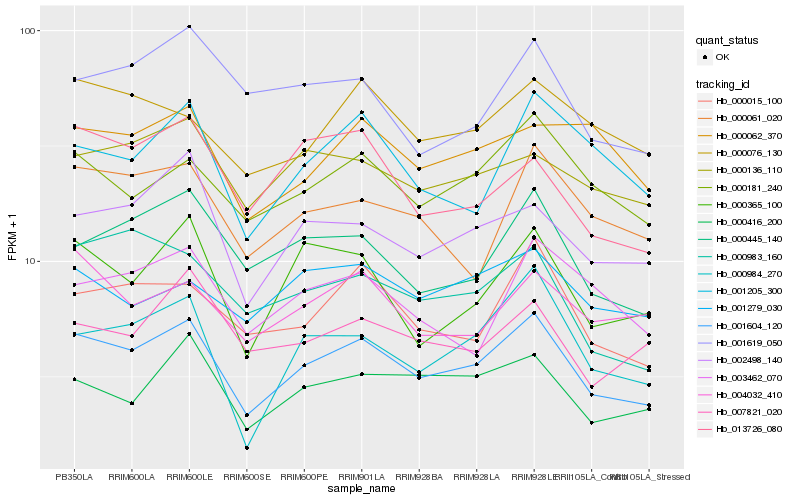
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001604_120 |
0.0 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000181_240 |
0.0929597304 |
- |
- |
PREDICTED: uncharacterized protein LOC105637672 [Jatropha curcas] |
| 3 |
Hb_002498_140 |
0.0950832508 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 4 |
Hb_000015_100 |
0.0953026204 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000445_140 |
0.0959976947 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 6 |
Hb_000416_200 |
0.0986844312 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 7 |
Hb_000365_100 |
0.098761546 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000984_270 |
0.1001822018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000136_110 |
0.1053852079 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 10 |
Hb_000061_020 |
0.1061682255 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 11 |
Hb_001205_300 |
0.1063108321 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 12 |
Hb_000983_160 |
0.1076367552 |
- |
- |
hypothetical protein B456_009G199800 [Gossypium raimondii] |
| 13 |
Hb_000076_130 |
0.1092008962 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 14 |
Hb_013726_080 |
0.1099545143 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 15 |
Hb_004032_410 |
0.1099662437 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007821_020 |
0.1116190811 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001619_050 |
0.1121715465 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001279_030 |
0.1124255738 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 19 |
Hb_003462_070 |
0.1132283667 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 20 |
Hb_000062_370 |
0.1145570787 |
- |
- |
beta-ketoacyl-ACP reductase [Vernicia fordii] |