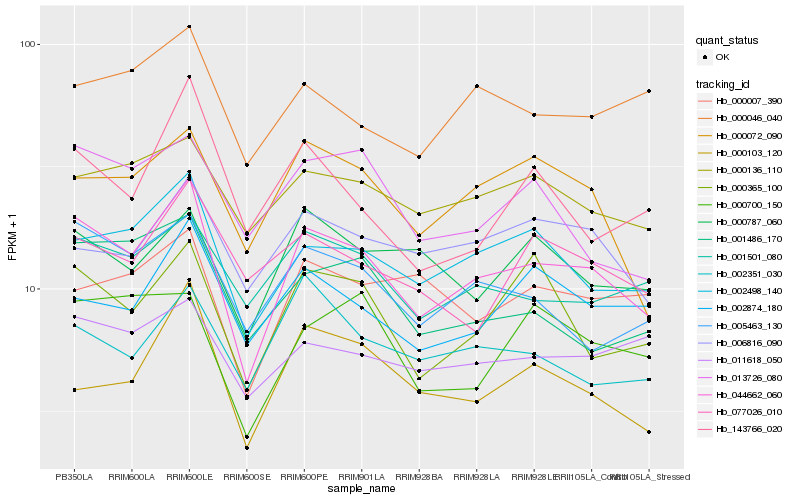
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044662_060 |
0.0 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002498_140 |
0.09624824 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 3 |
Hb_005463_130 |
0.1092242099 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 4 |
Hb_000365_100 |
0.1118564317 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001501_080 |
0.1149159929 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000072_090 |
0.1173340814 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 7 |
Hb_001486_170 |
0.1194124942 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 8 |
Hb_000136_110 |
0.1196210774 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 9 |
Hb_000103_120 |
0.1202313739 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002874_180 |
0.1223282834 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000787_060 |
0.1239958469 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_013726_080 |
0.1245669753 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 13 |
Hb_006816_090 |
0.1249713443 |
- |
- |
PREDICTED: uncharacterized protein LOC105647425 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002351_030 |
0.1260666647 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_011618_050 |
0.1261588051 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 16 |
Hb_000046_040 |
0.1279846481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000007_390 |
0.1284479898 |
- |
- |
hypothetical protein JCGZ_18250 [Jatropha curcas] |
| 18 |
Hb_000700_150 |
0.1286128061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_143766_020 |
0.1290144751 |
- |
- |
putative phospholipase A2 [Gossypium arboreum] |
| 20 |
Hb_077026_010 |
0.1296947384 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |