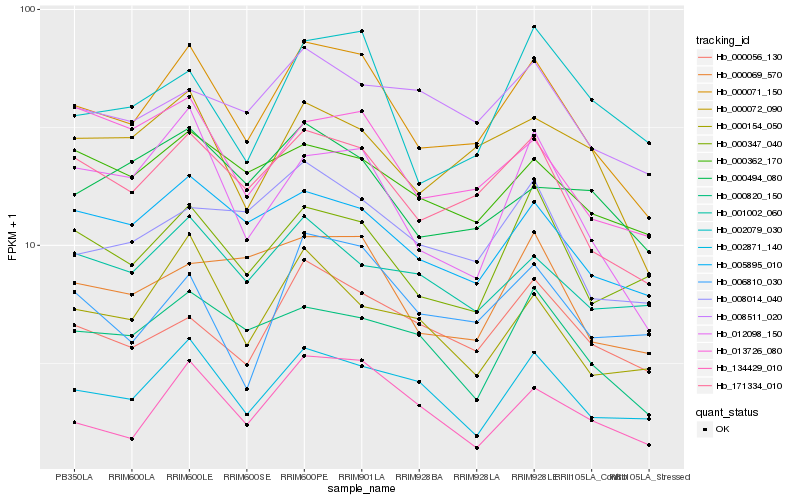
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000071_150 |
0.0 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 2 |
Hb_171334_010 |
0.0690956306 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 3 |
Hb_005895_010 |
0.0979836493 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002871_140 |
0.0998935907 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 5 |
Hb_000072_090 |
0.1006456065 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 6 |
Hb_012098_150 |
0.1042374817 |
- |
- |
ARC5 protein [Manihot esculenta] |
| 7 |
Hb_000056_130 |
0.1067929879 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 8 |
Hb_000347_040 |
0.1070150389 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 9 |
Hb_008014_040 |
0.1096813275 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 10 |
Hb_000820_150 |
0.1107576302 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 11 |
Hb_000069_570 |
0.112202226 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000362_170 |
0.1145008479 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 13 |
Hb_013726_080 |
0.114749215 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 14 |
Hb_134429_010 |
0.1159758132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002079_030 |
0.1160516868 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001002_060 |
0.1172069328 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 17 |
Hb_000494_080 |
0.1174508853 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_008511_020 |
0.1177853767 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 19 |
Hb_006810_030 |
0.1178755178 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000154_050 |
0.118079929 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |