| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
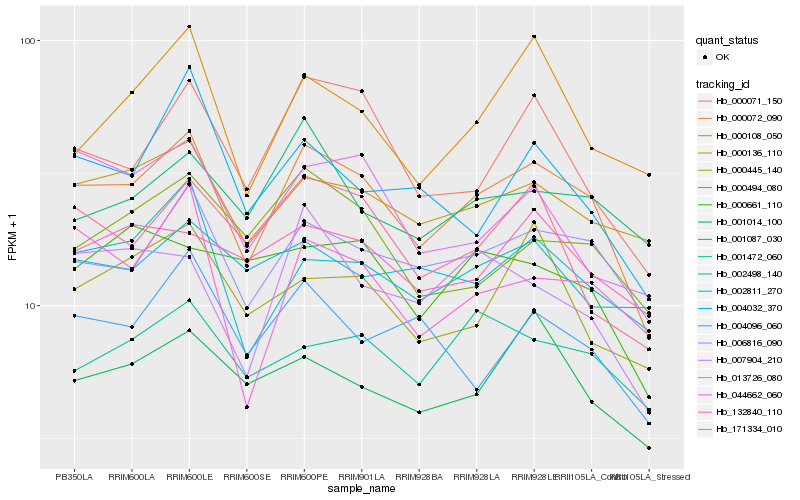
Hb_000072_090 |
0.0 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 2 |
Hb_006816_090 |
0.0847333382 |
- |
- |
PREDICTED: uncharacterized protein LOC105647425 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000071_150 |
0.1006456065 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 4 |
Hb_000136_110 |
0.102313471 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 5 |
Hb_171334_010 |
0.109002906 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 6 |
Hb_001472_060 |
0.1145053456 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001014_100 |
0.1147206889 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 8 |
Hb_000494_080 |
0.1155145981 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_007904_210 |
0.115587703 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1D [Jatropha curcas] |
| 10 |
Hb_013726_080 |
0.1169218592 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 11 |
Hb_000108_050 |
0.1170068559 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 12 |
Hb_044662_060 |
0.1173340814 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 13 |
Hb_132840_110 |
0.1174545537 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_002498_140 |
0.1175726082 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 15 |
Hb_002811_270 |
0.1182475171 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 16 |
Hb_000445_140 |
0.1191785949 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 17 |
Hb_001087_030 |
0.1193105826 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 18 |
Hb_004096_060 |
0.1213315277 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 19 |
Hb_004032_370 |
0.1220559187 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 20 |
Hb_000661_110 |
0.1237191619 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |