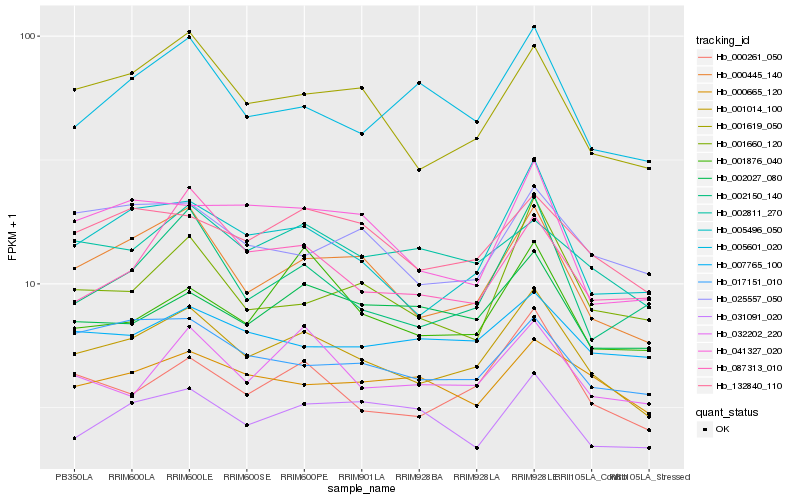
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001014_100 |
0.0 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 2 |
Hb_000445_140 |
0.0613544854 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 3 |
Hb_005496_050 |
0.0695335637 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 4 |
Hb_000261_050 |
0.0697395136 |
- |
- |
PREDICTED: probable magnesium transporter NIPA2 [Populus euphratica] |
| 5 |
Hb_001619_050 |
0.0722727605 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 6 |
Hb_132840_110 |
0.0749963172 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_002811_270 |
0.0843655476 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 8 |
Hb_032202_220 |
0.0873488504 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_017151_010 |
0.0889084471 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 10 |
Hb_002027_080 |
0.0905370592 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 11 |
Hb_005601_020 |
0.090860756 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_002150_140 |
0.0929036864 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 13 |
Hb_041327_020 |
0.0931563207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000665_120 |
0.0936115793 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 15 |
Hb_001660_120 |
0.0952513229 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_031091_020 |
0.0957086777 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 17 |
Hb_025557_050 |
0.0957559015 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 18 |
Hb_001876_040 |
0.096108094 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 19 |
Hb_087313_010 |
0.0969920381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007765_100 |
0.097089643 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |