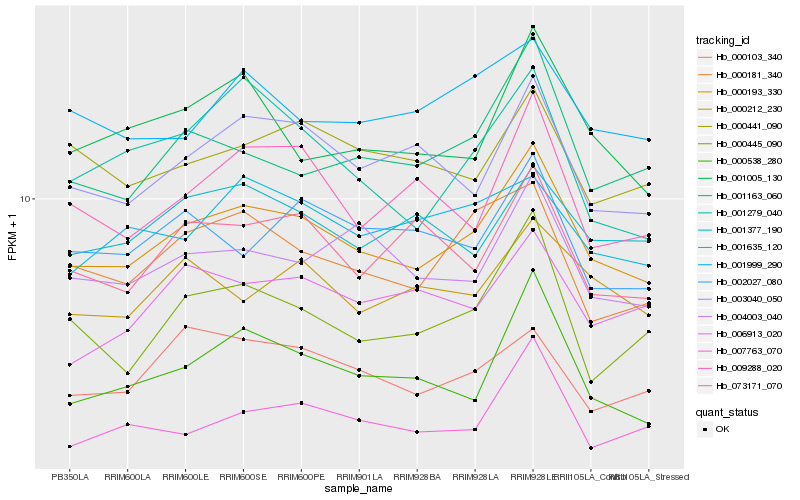
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000193_330 |
0.0 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 2 |
Hb_000538_280 |
0.0679690173 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 3 |
Hb_003040_050 |
0.068396428 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 4 |
Hb_000181_340 |
0.0684102552 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 5 |
Hb_001163_060 |
0.0699314699 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 6 |
Hb_004003_040 |
0.0700529306 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 7 |
Hb_007763_070 |
0.0712007741 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001005_130 |
0.0712768114 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 9 |
Hb_000445_090 |
0.072947148 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 10 |
Hb_000212_230 |
0.0760835591 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 11 |
Hb_001999_290 |
0.0764425475 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000103_340 |
0.0767782573 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 13 |
Hb_001377_190 |
0.0768571834 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_073171_070 |
0.0772634604 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 15 |
Hb_009288_020 |
0.0772903733 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 16 |
Hb_006913_020 |
0.0777876223 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000441_090 |
0.0782541102 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 18 |
Hb_002027_080 |
0.0790875394 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 19 |
Hb_001279_040 |
0.0792452348 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 20 |
Hb_001635_120 |
0.0793145973 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |