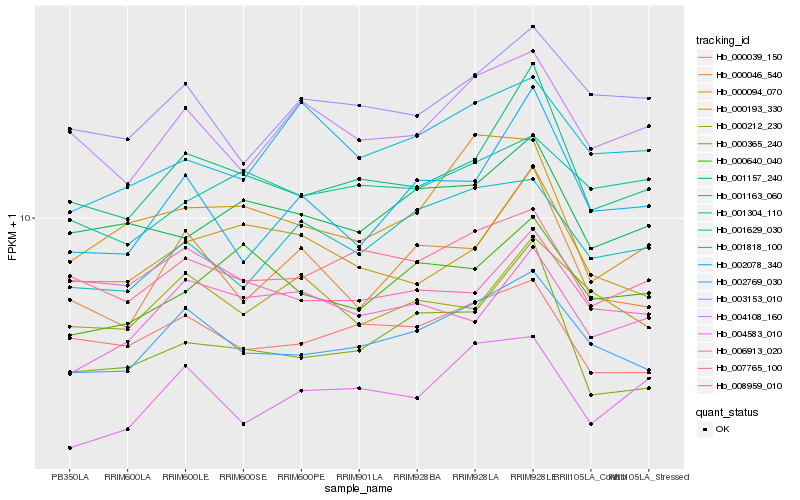
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002769_030 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000039_150 |
0.0644969226 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008959_010 |
0.0668107385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003153_010 |
0.0700505703 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 5 |
Hb_001163_060 |
0.0706646398 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 6 |
Hb_001629_030 |
0.0768498682 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 7 |
Hb_004108_160 |
0.0769840339 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 8 |
Hb_000046_540 |
0.0774953759 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 9 |
Hb_000212_230 |
0.0779370695 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 10 |
Hb_002078_340 |
0.0859653507 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007765_100 |
0.0863071101 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 12 |
Hb_001304_110 |
0.0871286447 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 13 |
Hb_001157_240 |
0.087945791 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 14 |
Hb_000094_070 |
0.0883170562 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_006913_020 |
0.0886288891 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000365_240 |
0.0888058395 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 17 |
Hb_004583_010 |
0.0892538168 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22150, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000640_040 |
0.0901921154 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000193_330 |
0.090468018 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 20 |
Hb_001818_100 |
0.090774483 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |