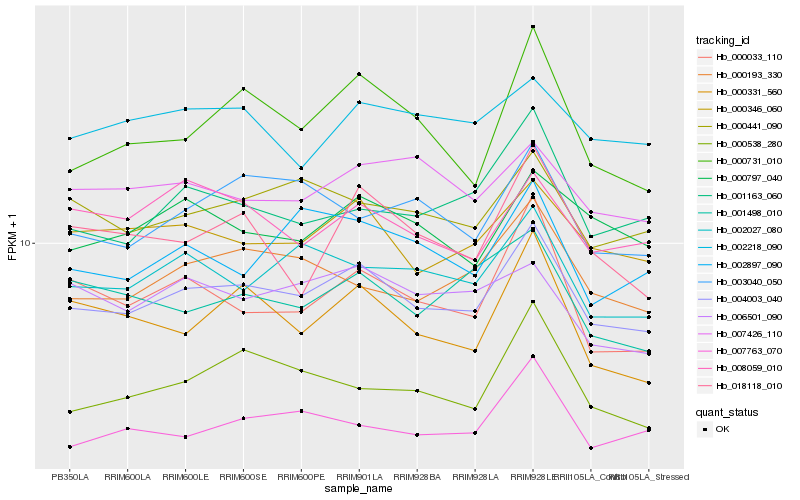
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004003_040 |
0.0 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 2 |
Hb_000346_060 |
0.0629592957 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 3 |
Hb_002027_080 |
0.068492997 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 4 |
Hb_000441_090 |
0.0690339211 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 5 |
Hb_000193_330 |
0.0700529306 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 6 |
Hb_000797_040 |
0.0745721128 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 7 |
Hb_000731_010 |
0.0765337545 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001163_060 |
0.0769165131 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 9 |
Hb_007763_070 |
0.0784830661 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003040_050 |
0.0791167649 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 11 |
Hb_002897_090 |
0.0792621123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007426_110 |
0.0802941978 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 13 |
Hb_000033_110 |
0.080335506 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 14 |
Hb_001498_010 |
0.0805133206 |
- |
- |
PREDICTED: uncharacterized protein LOC105648994 [Jatropha curcas] |
| 15 |
Hb_000331_560 |
0.0815958814 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 16 |
Hb_002218_090 |
0.0817755348 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 17 |
Hb_008059_010 |
0.0821637082 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 18 |
Hb_018118_010 |
0.0828032329 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 19 |
Hb_006501_090 |
0.0831121306 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 20 |
Hb_000538_280 |
0.0840016026 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |