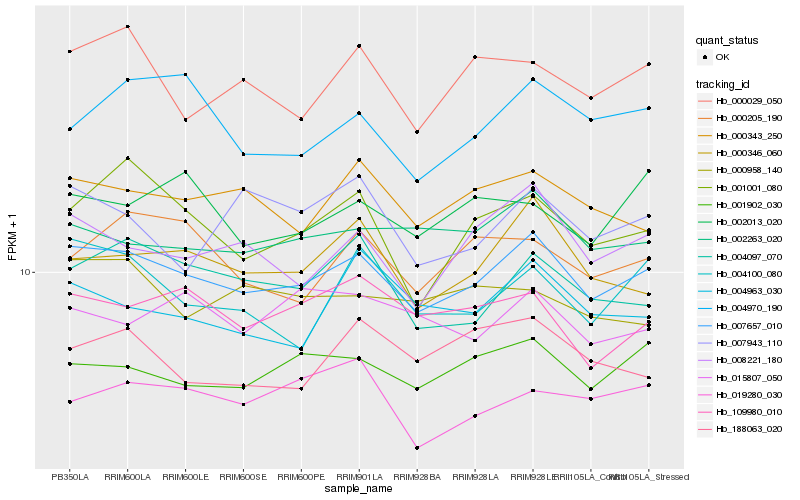
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007657_010 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_001902_030 |
0.057886615 |
- |
- |
PREDICTED: SKP1-like protein 21 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008221_180 |
0.0592717603 |
- |
- |
PREDICTED: uncharacterized protein LOC105644223 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000346_060 |
0.0600691854 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 5 |
Hb_002263_020 |
0.0610001491 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 6 |
Hb_000343_250 |
0.062144196 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 7 |
Hb_004097_070 |
0.0636137616 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 8 |
Hb_109980_010 |
0.0658522795 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001001_080 |
0.0674324803 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 10 |
Hb_000029_050 |
0.06833232 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 11 |
Hb_004100_080 |
0.0699546082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004970_190 |
0.0711663982 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_004963_030 |
0.0712751981 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |
| 14 |
Hb_000205_190 |
0.0724851244 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_019280_030 |
0.0726177134 |
- |
- |
- |
| 16 |
Hb_002013_020 |
0.0738083152 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 17 |
Hb_015807_050 |
0.0741294095 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 18 |
Hb_188063_020 |
0.0750612815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000958_140 |
0.0754151456 |
- |
- |
PREDICTED: uncharacterized protein LOC105628714 [Jatropha curcas] |
| 20 |
Hb_007943_110 |
0.0759076004 |
- |
- |
PREDICTED: basic salivary proline-rich protein 3 [Jatropha curcas] |