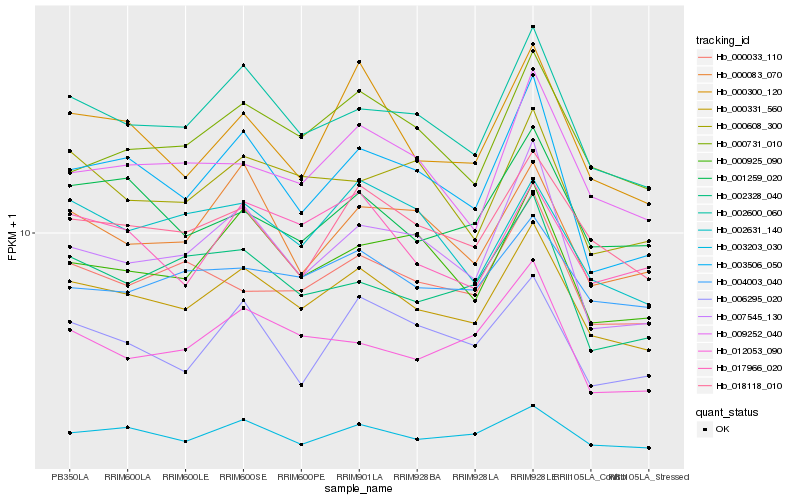
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_560 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 2 |
Hb_002600_060 |
0.0529647632 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 3 |
Hb_003203_030 |
0.0624285146 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 4 |
Hb_018118_010 |
0.0683943546 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 5 |
Hb_003506_050 |
0.077173194 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000300_120 |
0.0810828029 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009252_040 |
0.0811194692 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004003_040 |
0.0815958814 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_000731_010 |
0.0822253774 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000925_090 |
0.0826396342 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001259_020 |
0.0826947906 |
- |
- |
PREDICTED: translocase of chloroplast 90, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000033_110 |
0.0845604347 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 13 |
Hb_017966_020 |
0.0860208523 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03560, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000608_300 |
0.087391114 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 15 |
Hb_002328_040 |
0.0893360866 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000083_070 |
0.0909332747 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_012053_090 |
0.0910432471 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 18 |
Hb_007545_130 |
0.0917143352 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002631_140 |
0.0919450119 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 20 |
Hb_006295_020 |
0.0928735469 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR isoform X1 [Jatropha curcas] |