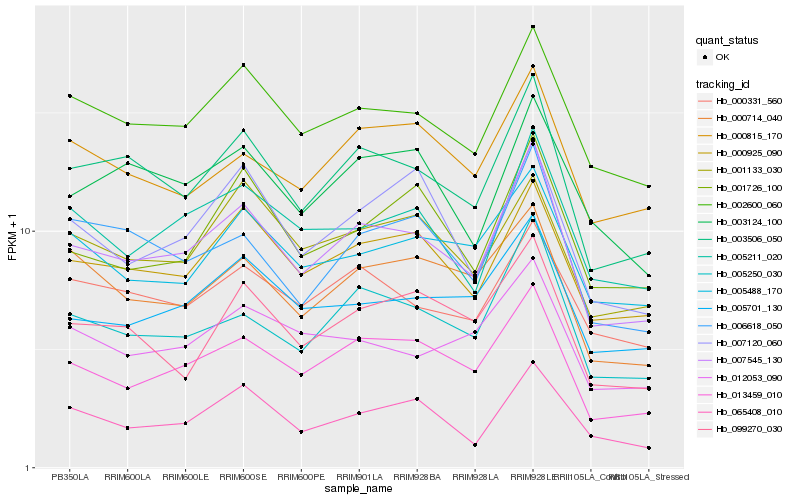
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007545_130 |
0.0 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003506_050 |
0.0551158428 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_013459_010 |
0.0558634876 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 4 |
Hb_005250_030 |
0.0671460082 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 5 |
Hb_000925_090 |
0.0727864195 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002600_060 |
0.0737749784 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 7 |
Hb_005211_020 |
0.0816252982 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_005701_130 |
0.0850059833 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 9 |
Hb_006618_050 |
0.0889075865 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_065408_010 |
0.0914789583 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000331_560 |
0.0917143352 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 12 |
Hb_001726_100 |
0.0929553992 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 13 |
Hb_099270_030 |
0.0951589875 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_005488_170 |
0.0951691032 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 15 |
Hb_003124_100 |
0.0958411382 |
- |
- |
- |
| 16 |
Hb_000815_170 |
0.0961446713 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 17 |
Hb_001133_030 |
0.096643393 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 18 |
Hb_000714_040 |
0.0973645939 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 19 |
Hb_012053_090 |
0.0977396406 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 20 |
Hb_007120_060 |
0.0988899559 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |