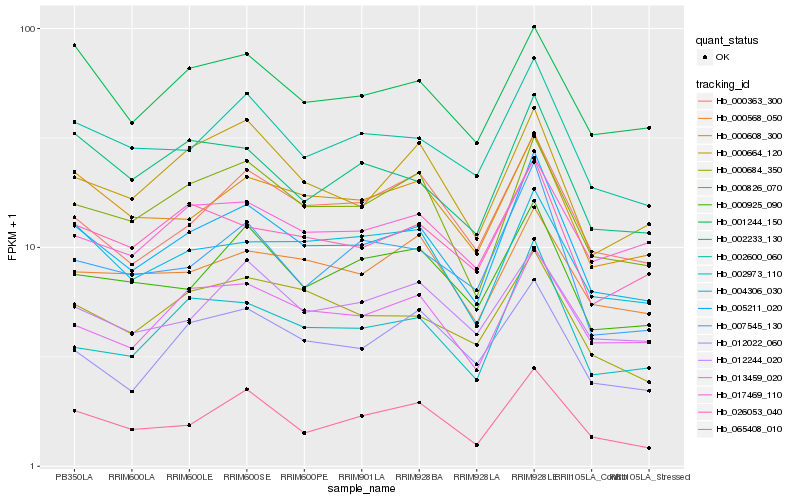
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005211_020 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002600_060 |
0.0667850467 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 3 |
Hb_000826_070 |
0.069989394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002973_110 |
0.0747020576 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_000608_300 |
0.0749517768 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 6 |
Hb_013459_020 |
0.0758988276 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 7 |
Hb_000925_090 |
0.0805028011 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000363_300 |
0.0808613052 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_065408_010 |
0.0814684379 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 10 |
Hb_007545_130 |
0.0816252982 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_026053_040 |
0.0826119324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004306_030 |
0.0837599919 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 13 |
Hb_000684_350 |
0.0838125207 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002233_130 |
0.086344189 |
- |
- |
transporter, putative [Ricinus communis] |
| 15 |
Hb_001244_150 |
0.0870519953 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 16 |
Hb_000664_120 |
0.088329554 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_017469_110 |
0.0886140759 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 18 |
Hb_012244_020 |
0.0887210063 |
- |
- |
calpain, putative [Ricinus communis] |
| 19 |
Hb_000568_050 |
0.08914434 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_012022_060 |
0.0912001777 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |