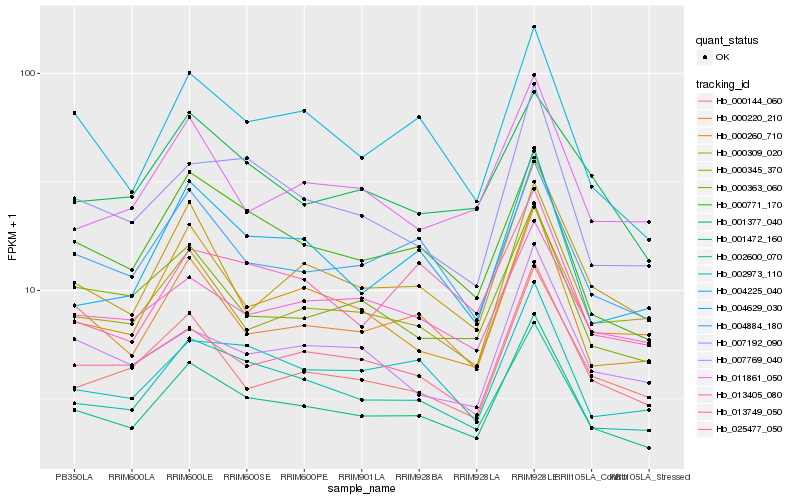
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001472_160 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 2 |
Hb_025477_050 |
0.0661830723 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_004884_180 |
0.0768025251 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000220_210 |
0.0774601375 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004225_040 |
0.0807307192 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 6 |
Hb_000309_020 |
0.0808805234 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 7 |
Hb_002973_110 |
0.0815979954 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000771_170 |
0.083429311 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_007192_090 |
0.084321281 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 10 |
Hb_000144_060 |
0.0863219953 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000345_370 |
0.0875805134 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 12 |
Hb_000363_060 |
0.088294374 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000260_710 |
0.0893532499 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002600_070 |
0.0896170426 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 15 |
Hb_013405_080 |
0.0897211204 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 16 |
Hb_004629_030 |
0.0920303735 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_007769_040 |
0.092782068 |
- |
- |
PREDICTED: probable threonine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 18 |
Hb_011861_050 |
0.0928777269 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_013749_050 |
0.0958029334 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001377_040 |
0.0970914862 |
- |
- |
chitinase, putative [Ricinus communis] |