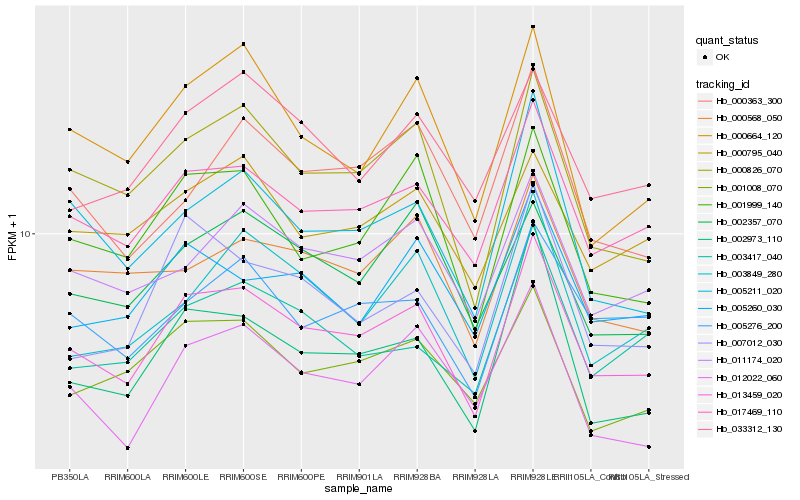
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013459_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 2 |
Hb_017469_110 |
0.0495022791 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 3 |
Hb_000826_070 |
0.0620955615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011174_020 |
0.0704707915 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 5 |
Hb_002973_110 |
0.0714636303 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_001999_140 |
0.071477713 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_001008_070 |
0.0753091456 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 8 |
Hb_005276_200 |
0.075782512 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005211_020 |
0.0758988276 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_012022_060 |
0.0774245391 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005260_030 |
0.0801094725 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 12 |
Hb_000795_040 |
0.0803028553 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 13 |
Hb_007012_030 |
0.080484568 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000664_120 |
0.0805166809 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003849_280 |
0.0807405532 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 16 |
Hb_000568_050 |
0.081193204 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003417_040 |
0.0813814862 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 18 |
Hb_033312_130 |
0.0819677545 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002357_070 |
0.0821301023 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 20 |
Hb_000363_300 |
0.0827786041 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |