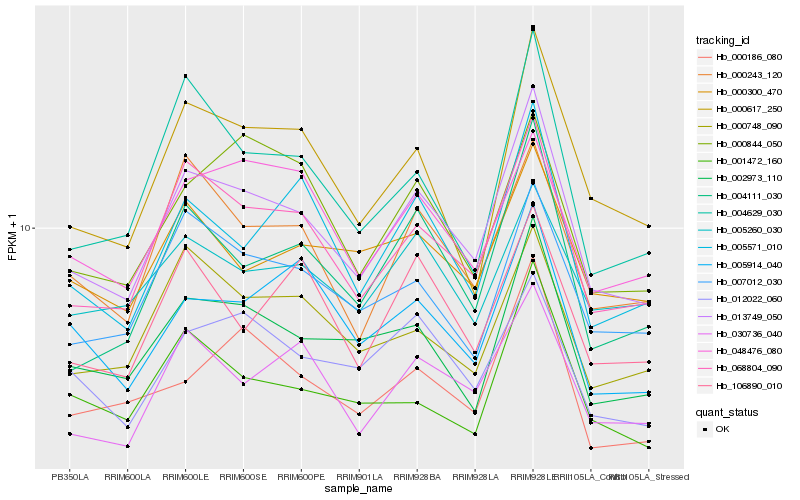
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013749_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 2 |
Hb_068804_090 |
0.0541082933 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 3 |
Hb_002973_110 |
0.0792076952 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_004111_030 |
0.0813945208 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_030736_040 |
0.0833132183 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000243_120 |
0.0845678855 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000617_250 |
0.0855247393 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005571_010 |
0.0862582683 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 9 |
Hb_007012_030 |
0.086330731 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_012022_060 |
0.0907960054 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004629_030 |
0.0925335163 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_000748_090 |
0.092810118 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 13 |
Hb_000186_080 |
0.0947165266 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 14 |
Hb_005914_040 |
0.0957076618 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 15 |
Hb_001472_160 |
0.0958029334 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 16 |
Hb_000300_470 |
0.0976473532 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 17 |
Hb_048476_080 |
0.0983162615 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 18 |
Hb_106890_010 |
0.1002830639 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_005260_030 |
0.100342495 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 20 |
Hb_000844_050 |
0.1005360978 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |