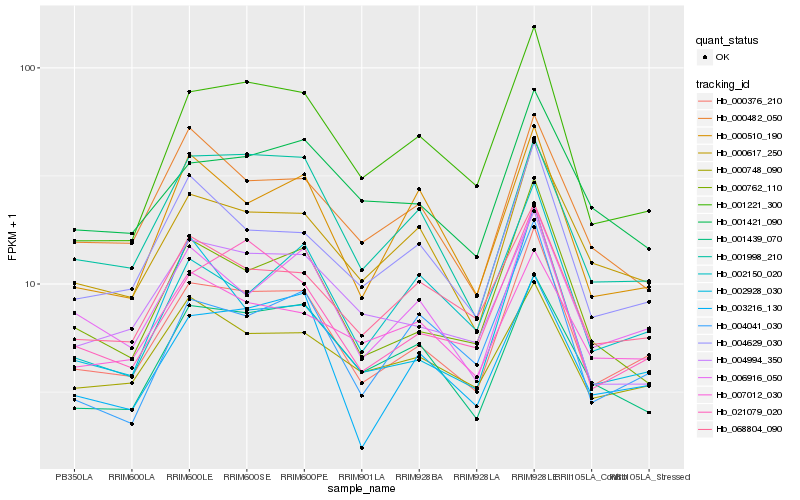
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000376_210 |
0.0 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 2 |
Hb_006916_050 |
0.08271286 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_000617_250 |
0.0829394944 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_021079_020 |
0.0850382439 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 5 |
Hb_004629_030 |
0.0874369748 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_000762_110 |
0.091063631 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_068804_090 |
0.0928159175 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 8 |
Hb_001221_300 |
0.0935204525 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003216_130 |
0.099983065 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 10 |
Hb_001998_210 |
0.1025072667 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_000748_090 |
0.1028237827 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 12 |
Hb_001421_090 |
0.1030922898 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 13 |
Hb_001439_070 |
0.105208747 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000510_190 |
0.1055128597 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 15 |
Hb_002150_020 |
0.1066505752 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_007012_030 |
0.1079375137 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_004994_350 |
0.1087053774 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000482_050 |
0.1089968017 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004041_030 |
0.11018128 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 20 |
Hb_002928_030 |
0.1122874452 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |