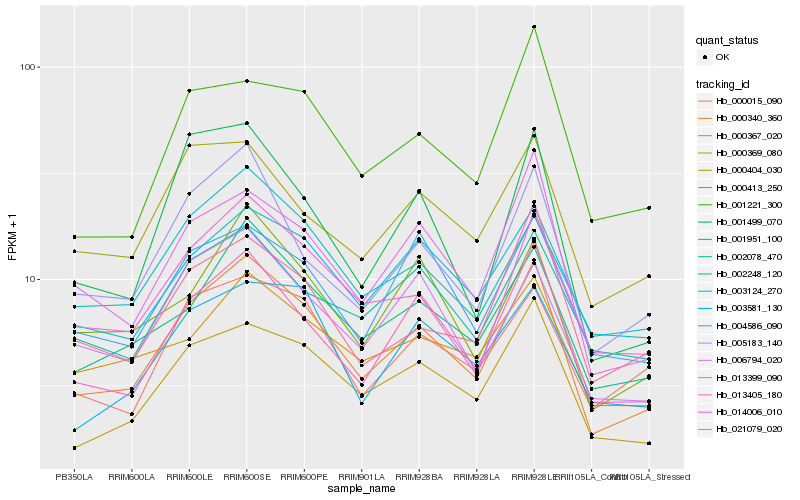
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_360 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 2 |
Hb_014006_010 |
0.0770569554 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 3 |
Hb_000369_080 |
0.0844695292 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 4 |
Hb_001951_100 |
0.0866105264 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 5 |
Hb_013405_180 |
0.0920717947 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 6 |
Hb_013399_090 |
0.0929879938 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_005183_140 |
0.0946490305 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 8 |
Hb_002248_120 |
0.1013615077 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 9 |
Hb_006794_020 |
0.1033227929 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 10 |
Hb_000413_250 |
0.1045929761 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 11 |
Hb_003124_270 |
0.1065929105 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 12 |
Hb_021079_020 |
0.1066793273 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 13 |
Hb_000015_090 |
0.1071514621 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 14 |
Hb_001499_070 |
0.1079852244 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 15 |
Hb_002078_470 |
0.1087358537 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003581_130 |
0.1100072554 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000404_030 |
0.1115329861 |
- |
- |
PREDICTED: protein NAP1 [Jatropha curcas] |
| 18 |
Hb_004586_090 |
0.1122080966 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001221_300 |
0.1124438381 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000367_020 |
0.1130953752 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |