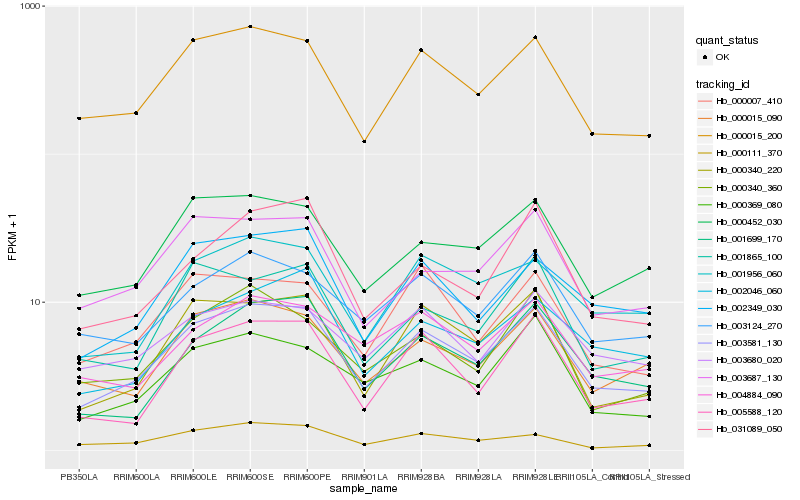
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003581_130 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000015_200 |
0.0732764377 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 3 |
Hb_000452_030 |
0.0941367228 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 4 |
Hb_000015_090 |
0.0946061667 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 5 |
Hb_000340_220 |
0.0951033063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002349_030 |
0.0981284973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000369_080 |
0.0988166076 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 8 |
Hb_001699_170 |
0.0988716201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003124_270 |
0.0994564373 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 10 |
Hb_003687_130 |
0.0995391809 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 11 |
Hb_005588_120 |
0.1000002535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001956_060 |
0.1008453769 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 13 |
Hb_001865_100 |
0.1068070979 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 14 |
Hb_003680_020 |
0.1083377222 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_004884_090 |
0.1083454943 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 16 |
Hb_000111_370 |
0.1084553113 |
- |
- |
PREDICTED: cation/H(+) antiporter 2-like [Jatropha curcas] |
| 17 |
Hb_031089_050 |
0.1092528529 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_002046_060 |
0.1098290639 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 19 |
Hb_000340_360 |
0.1100072554 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 20 |
Hb_000007_410 |
0.1101003942 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |