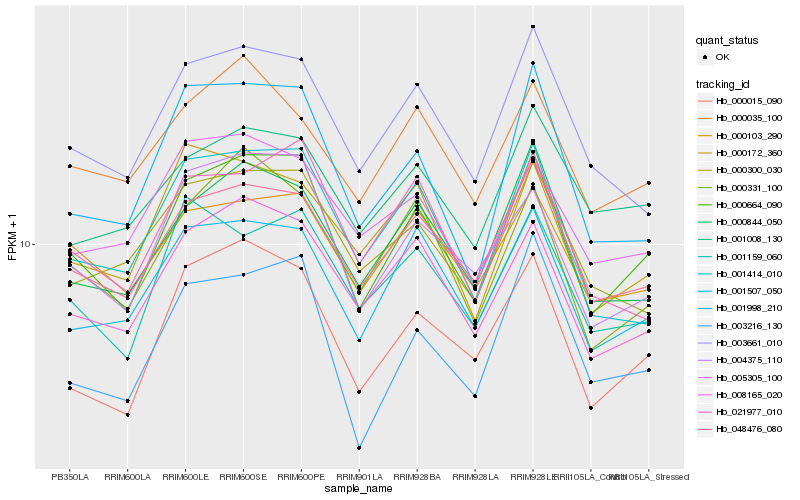
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000664_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 2 |
Hb_004375_110 |
0.0724244114 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 3 |
Hb_048476_080 |
0.0741162681 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 4 |
Hb_008165_020 |
0.0763707607 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000015_090 |
0.0776556371 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 6 |
Hb_001507_050 |
0.0788456961 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000844_050 |
0.0828713137 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 8 |
Hb_001998_210 |
0.0851678437 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_000331_100 |
0.0851784971 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003661_010 |
0.0863069729 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001159_060 |
0.0886627653 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 12 |
Hb_001414_010 |
0.0886682018 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 13 |
Hb_000103_290 |
0.0904056597 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 14 |
Hb_021977_010 |
0.0925030794 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 15 |
Hb_000300_030 |
0.0956306389 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 16 |
Hb_003216_130 |
0.0964605144 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 17 |
Hb_000035_100 |
0.0964612253 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000172_360 |
0.0980412775 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 19 |
Hb_001008_130 |
0.0991869893 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 20 |
Hb_005305_100 |
0.099242869 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |