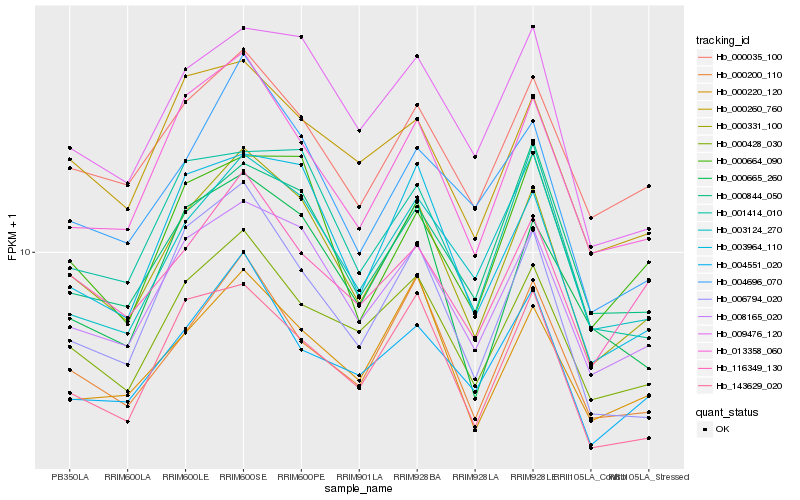
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 2 |
Hb_008165_020 |
0.0454601142 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000428_030 |
0.0640484357 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 4 |
Hb_013358_060 |
0.0689181361 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 5 |
Hb_000035_100 |
0.0718377367 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_003964_110 |
0.0734095307 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 7 |
Hb_009476_120 |
0.0777060614 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 8 |
Hb_000200_110 |
0.0793745997 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004696_070 |
0.0800203164 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 10 |
Hb_143629_020 |
0.0802393124 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 11 |
Hb_000260_760 |
0.0806279547 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 12 |
Hb_001414_010 |
0.0808959764 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 13 |
Hb_006794_020 |
0.0819261387 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 14 |
Hb_000665_260 |
0.0840245543 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000220_120 |
0.0848114959 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000664_090 |
0.0851784971 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 17 |
Hb_003124_270 |
0.0852290332 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 18 |
Hb_000844_050 |
0.0861769061 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 19 |
Hb_004551_020 |
0.0863197388 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 20 |
Hb_116349_130 |
0.0863625764 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |