| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
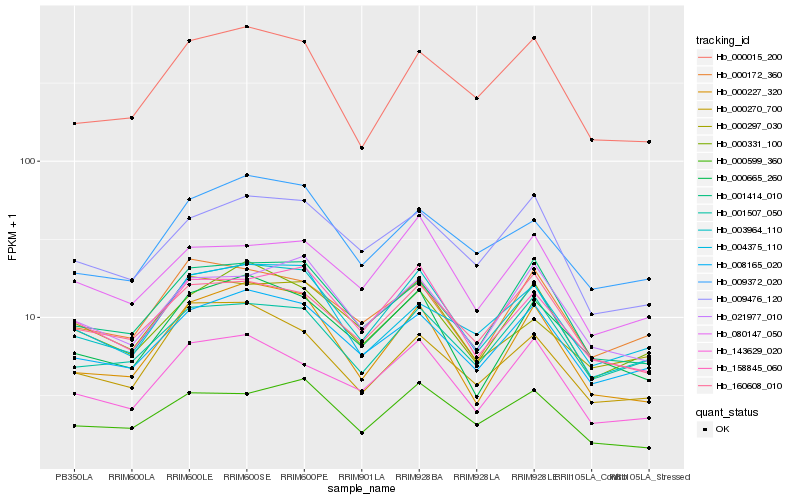
Hb_003964_110 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 2 |
Hb_158845_060 |
0.0637228286 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008165_020 |
0.0667355175 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000227_320 |
0.0728285099 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000331_100 |
0.0734095307 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001414_010 |
0.0747520376 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 7 |
Hb_000665_260 |
0.076837266 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000599_360 |
0.0781631699 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_009372_020 |
0.0787075306 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 10 |
Hb_143629_020 |
0.081539662 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 11 |
Hb_009476_120 |
0.0822276577 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 12 |
Hb_000015_200 |
0.0846534596 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 13 |
Hb_021977_010 |
0.0865967405 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 14 |
Hb_001507_050 |
0.0880830865 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_080147_050 |
0.0907709554 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 16 |
Hb_000270_700 |
0.0911118945 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 17 |
Hb_000172_360 |
0.091142906 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 18 |
Hb_000297_030 |
0.0916162161 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_160608_010 |
0.0916338016 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_004375_110 |
0.0916611654 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |