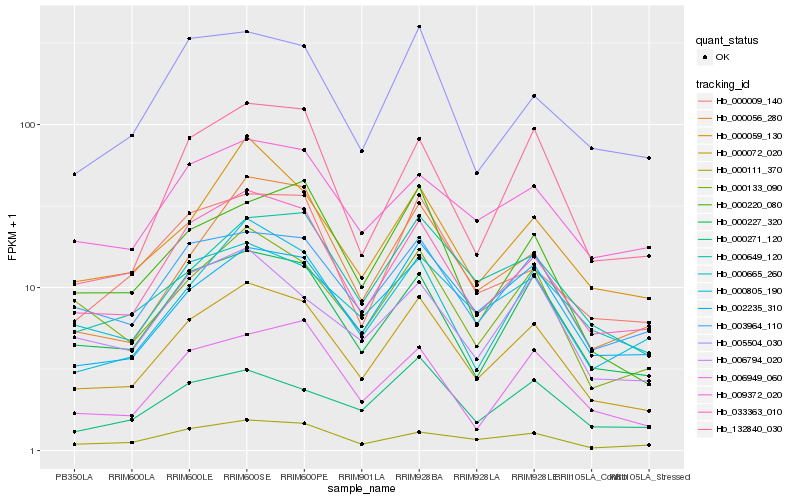
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_020 |
0.0 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 2 |
Hb_033363_010 |
0.0650845397 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 3 |
Hb_000227_320 |
0.081529278 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_003964_110 |
0.0938754967 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002235_310 |
0.0953683836 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 6 |
Hb_000805_190 |
0.0965155352 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 7 |
Hb_000133_090 |
0.0989560494 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 8 |
Hb_000649_120 |
0.1005091394 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000111_370 |
0.1060581944 |
- |
- |
PREDICTED: cation/H(+) antiporter 2-like [Jatropha curcas] |
| 10 |
Hb_000059_130 |
0.1065398173 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000056_280 |
0.1090690276 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 12 |
Hb_005504_030 |
0.110280595 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 13 |
Hb_000220_080 |
0.1103719797 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_000009_140 |
0.1118758631 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 15 |
Hb_006949_060 |
0.1131895348 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 16 |
Hb_132840_030 |
0.1134847424 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 17 |
Hb_009372_020 |
0.113682245 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 18 |
Hb_006794_020 |
0.1154980617 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 19 |
Hb_000665_260 |
0.1158290592 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000271_120 |
0.1164932891 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |