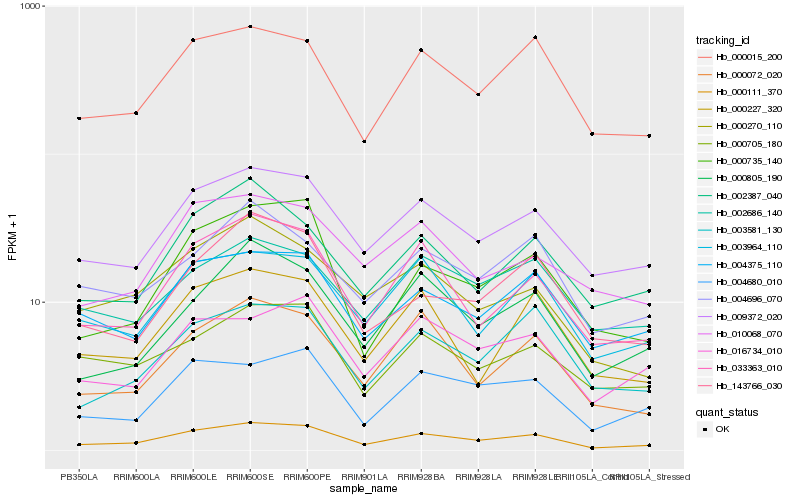
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_370 |
0.0 |
- |
- |
PREDICTED: cation/H(+) antiporter 2-like [Jatropha curcas] |
| 2 |
Hb_009372_020 |
0.0751213066 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 3 |
Hb_033363_010 |
0.0904445774 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 4 |
Hb_000015_200 |
0.0928384065 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 5 |
Hb_004375_110 |
0.096530238 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 6 |
Hb_003964_110 |
0.1020483764 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 7 |
Hb_004680_010 |
0.1032076796 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000072_020 |
0.1060581944 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 9 |
Hb_016734_010 |
0.1076872521 |
- |
- |
PREDICTED: probable DNA-3-methyladenine glycosylase 2 [Jatropha curcas] |
| 10 |
Hb_003581_130 |
0.1084553113 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000805_190 |
0.1097848368 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 12 |
Hb_002387_040 |
0.1109290632 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 13 |
Hb_000735_140 |
0.1113755841 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 14 |
Hb_000705_180 |
0.1116009063 |
- |
- |
hypothetical protein JCGZ_02923 [Jatropha curcas] |
| 15 |
Hb_004696_070 |
0.1133629347 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 16 |
Hb_000227_320 |
0.1136723185 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_143766_030 |
0.1148363076 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_002686_140 |
0.1150240465 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000270_110 |
0.1151038857 |
- |
- |
PREDICTED: calcium-dependent protein kinase 10 [Jatropha curcas] |
| 20 |
Hb_010068_070 |
0.1153079511 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |