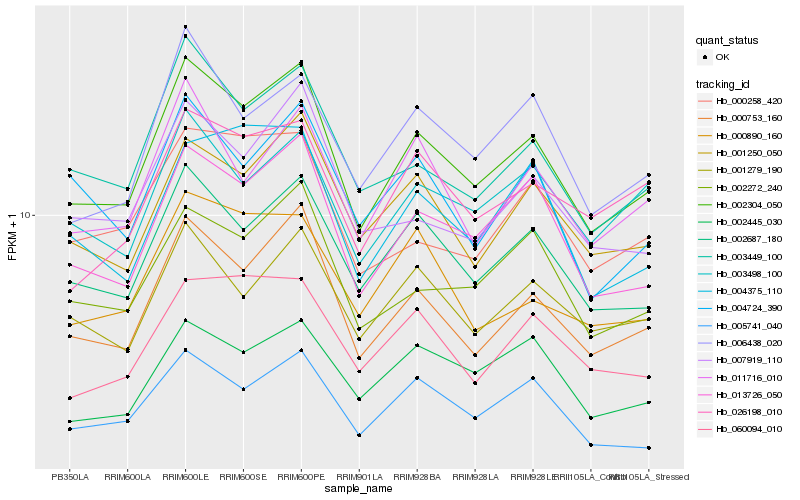
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002304_050 |
0.0 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 2 |
Hb_000753_160 |
0.0510283292 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 3 |
Hb_013726_050 |
0.0610535548 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 4 |
Hb_002687_180 |
0.0617005785 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 5 |
Hb_003449_100 |
0.0723503141 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 6 |
Hb_001250_050 |
0.0756365354 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002272_240 |
0.0783900716 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_006438_020 |
0.0810763619 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 9 |
Hb_011716_010 |
0.0835291246 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 10 |
Hb_001279_190 |
0.0842807186 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005741_040 |
0.0857995861 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 12 |
Hb_007919_110 |
0.0869361975 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 13 |
Hb_000258_420 |
0.0905215948 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 14 |
Hb_003498_100 |
0.0922728538 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 15 |
Hb_002445_030 |
0.0927275268 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 16 |
Hb_060094_010 |
0.0933241337 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004724_390 |
0.0933809016 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 18 |
Hb_004375_110 |
0.0938305042 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 19 |
Hb_000890_160 |
0.0939016958 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 20 |
Hb_026198_010 |
0.0941201089 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |