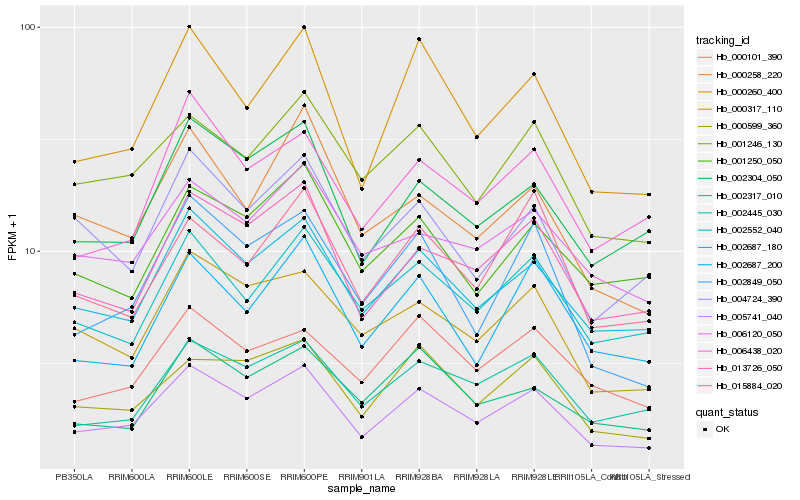
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005741_040 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 2 |
Hb_000260_400 |
0.0629127784 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 3 |
Hb_002687_180 |
0.0668931033 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 4 |
Hb_013726_050 |
0.0705863805 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 5 |
Hb_002849_050 |
0.0784553045 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 6 |
Hb_000599_360 |
0.080283883 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 7 |
Hb_000317_110 |
0.0843214247 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 8 |
Hb_002304_050 |
0.0857995861 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 9 |
Hb_002445_030 |
0.0864590006 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 10 |
Hb_001246_130 |
0.0874131792 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 11 |
Hb_002552_040 |
0.0915593727 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 12 |
Hb_002687_200 |
0.0956542794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006438_020 |
0.0964867996 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 14 |
Hb_000101_390 |
0.096759716 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 15 |
Hb_002317_010 |
0.0975299968 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_004724_390 |
0.0999550411 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 17 |
Hb_006120_050 |
0.1000234451 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 18 |
Hb_000258_220 |
0.1008839209 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_015884_020 |
0.1009305626 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 20 |
Hb_001250_050 |
0.1024751688 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |