| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
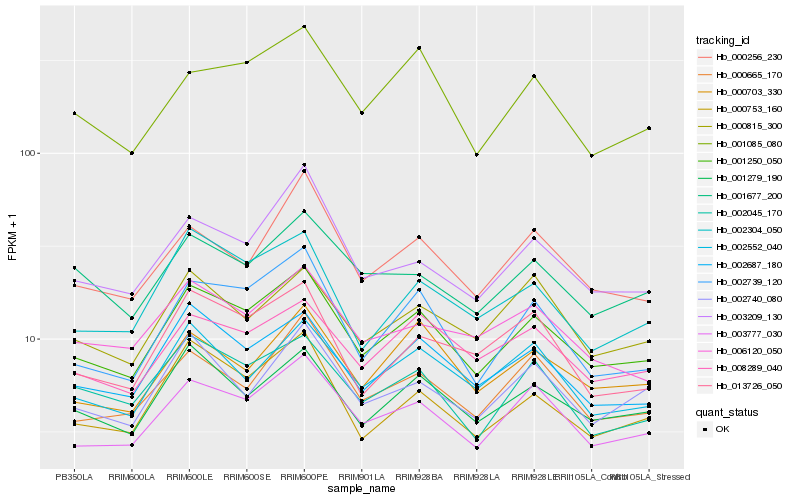
Hb_001250_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003777_030 |
0.0533254903 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 3 |
Hb_002687_180 |
0.0612305984 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 4 |
Hb_001279_190 |
0.062916398 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000753_160 |
0.0682426932 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 6 |
Hb_002739_120 |
0.068485757 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 7 |
Hb_008289_040 |
0.0714108714 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 8 |
Hb_002304_050 |
0.0756365354 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 9 |
Hb_002045_170 |
0.0761794019 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 10 |
Hb_000665_170 |
0.0766905118 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 11 |
Hb_002552_040 |
0.0784707749 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 12 |
Hb_001085_080 |
0.079446136 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 13 |
Hb_006120_050 |
0.0819530989 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 14 |
Hb_003209_130 |
0.0820357726 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 15 |
Hb_000703_330 |
0.0822154468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000256_230 |
0.0831013254 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 17 |
Hb_001677_200 |
0.0840520319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_013726_050 |
0.0842019326 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 19 |
Hb_002740_080 |
0.0844407088 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 20 |
Hb_000815_300 |
0.0845914994 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |