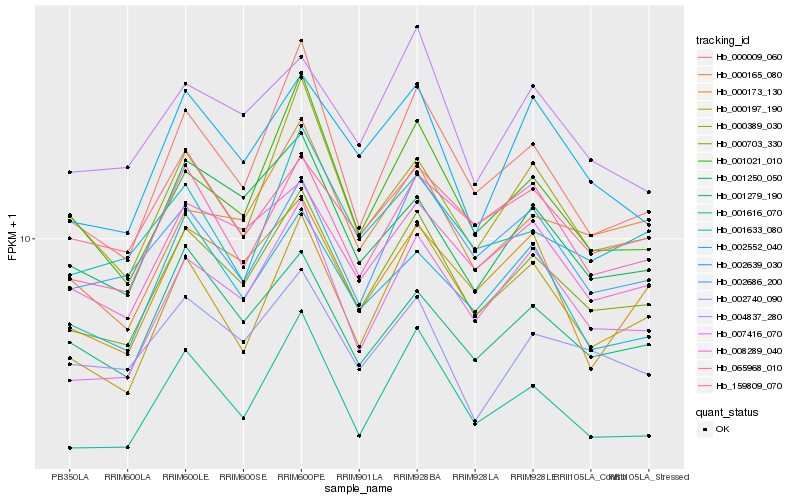
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_330 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007416_070 |
0.06538856 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 3 |
Hb_008289_040 |
0.0660388843 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 4 |
Hb_000009_060 |
0.0712407244 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 5 |
Hb_000197_190 |
0.0767858067 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_065968_010 |
0.0784963486 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002639_030 |
0.078671286 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 8 |
Hb_000165_080 |
0.0797507579 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002686_200 |
0.080628344 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 10 |
Hb_001021_010 |
0.0813563426 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 11 |
Hb_001250_050 |
0.0822154468 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001279_190 |
0.082232536 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002552_040 |
0.0827107804 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 14 |
Hb_000173_130 |
0.0828212323 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004837_280 |
0.0834687084 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 16 |
Hb_001616_070 |
0.0846374494 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 17 |
Hb_002740_090 |
0.0849744486 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 18 |
Hb_000389_030 |
0.0858941341 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 19 |
Hb_001633_080 |
0.0878046772 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 20 |
Hb_159809_070 |
0.0892534212 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |