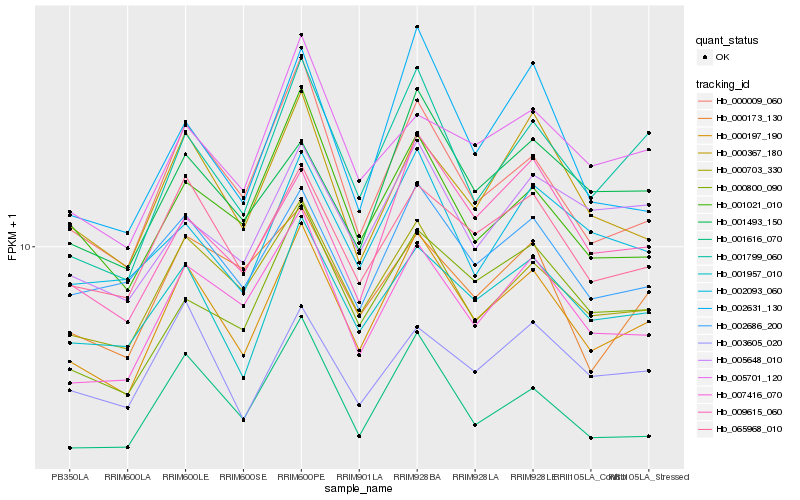
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000197_190 |
0.0 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_001799_060 |
0.0749391644 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 3 |
Hb_065968_010 |
0.0764982675 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000703_330 |
0.0767858067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001957_010 |
0.0778468328 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000009_060 |
0.0784312055 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 7 |
Hb_007416_070 |
0.0785551308 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 8 |
Hb_001616_070 |
0.0855224108 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 9 |
Hb_002686_200 |
0.0871759127 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 10 |
Hb_000367_180 |
0.091391641 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 11 |
Hb_000800_090 |
0.0928703151 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 12 |
Hb_009615_060 |
0.0967966446 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 13 |
Hb_002093_060 |
0.0978059984 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 14 |
Hb_002631_130 |
0.0984178309 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 15 |
Hb_001493_150 |
0.0988397875 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 16 |
Hb_003605_020 |
0.1013449444 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 17 |
Hb_001021_010 |
0.1016750649 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 18 |
Hb_005701_120 |
0.1025730001 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 19 |
Hb_005648_010 |
0.103076006 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 20 |
Hb_000173_130 |
0.1036256367 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |