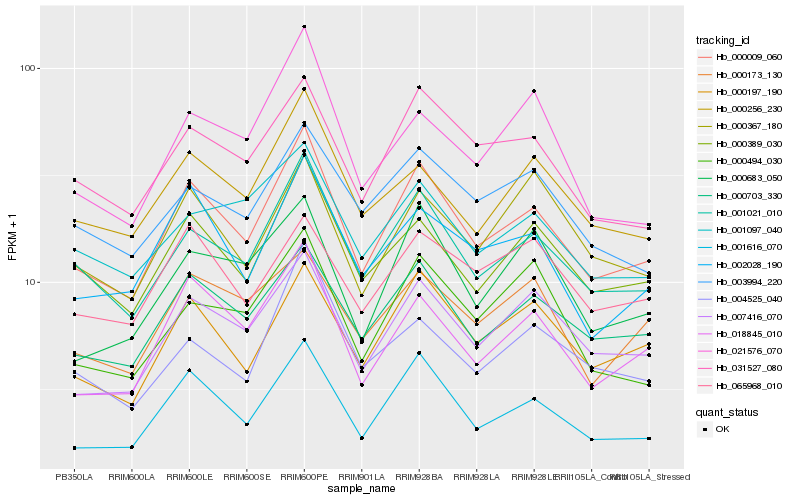
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_060 |
0.0 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 2 |
Hb_001616_070 |
0.0523545335 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 3 |
Hb_001021_010 |
0.0585296897 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 4 |
Hb_000703_330 |
0.0712407244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_018845_010 |
0.0757034684 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 6 |
Hb_007416_070 |
0.0766370667 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 7 |
Hb_000389_030 |
0.0781831619 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 8 |
Hb_000197_190 |
0.0784312055 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_002028_190 |
0.0853239442 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7-like [Jatropha curcas] |
| 10 |
Hb_031527_080 |
0.0857363701 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000256_230 |
0.0919302979 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 12 |
Hb_001097_040 |
0.1019782103 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 13 |
Hb_000683_050 |
0.1020720387 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 14 |
Hb_003994_220 |
0.1021686523 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 15 |
Hb_000173_130 |
0.1047095827 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_021576_070 |
0.1048378539 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 17 |
Hb_065968_010 |
0.1048470199 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000494_030 |
0.105245483 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 19 |
Hb_004525_040 |
0.105356901 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 20 |
Hb_000367_180 |
0.1058062154 |
- |
- |
Heparanase-2, putative [Ricinus communis] |