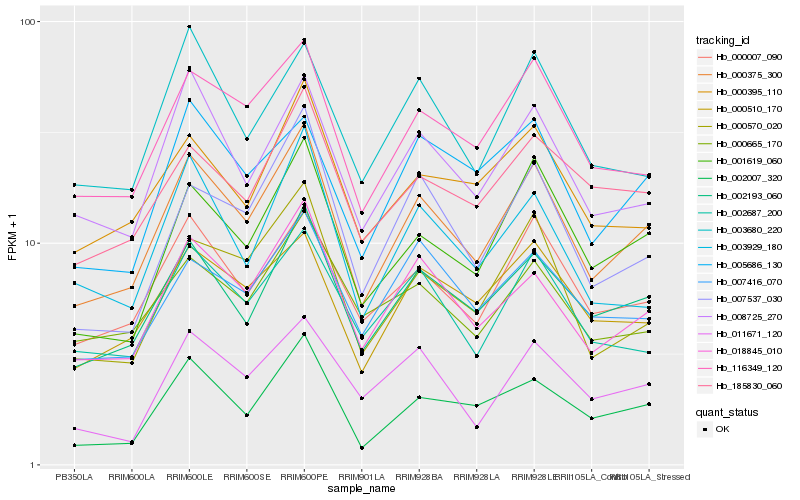
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000375_300 |
0.0 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 2 |
Hb_002193_060 |
0.0694840683 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 3 |
Hb_018845_010 |
0.0705198058 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 4 |
Hb_000007_090 |
0.0801946858 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 5 |
Hb_001619_060 |
0.080642852 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 6 |
Hb_116349_120 |
0.0820838999 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_008725_270 |
0.0879920854 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 8 |
Hb_000570_020 |
0.0894332372 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 9 |
Hb_000510_170 |
0.0929439546 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 10 |
Hb_002007_320 |
0.0930872045 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 11 |
Hb_002687_200 |
0.0972791134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007537_030 |
0.0982618042 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_185830_060 |
0.0996942277 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 14 |
Hb_000395_110 |
0.1003947392 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 15 |
Hb_007416_070 |
0.1037841291 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 16 |
Hb_005686_130 |
0.1041700591 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 17 |
Hb_003680_220 |
0.1043854608 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 18 |
Hb_000665_170 |
0.1057583086 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 19 |
Hb_003929_180 |
0.1079407628 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 20 |
Hb_011671_120 |
0.1084438251 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |