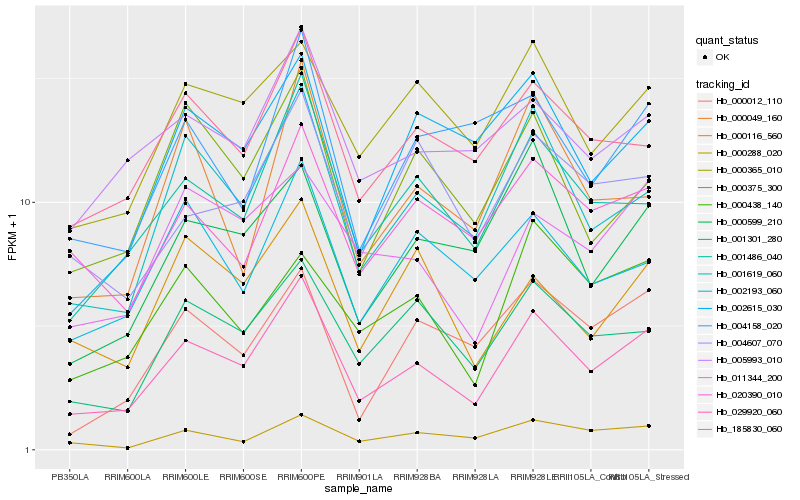
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029920_060 |
0.0 |
- |
- |
PREDICTED: UPF0554 protein-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001619_060 |
0.0933289214 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001486_040 |
0.1009192797 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 4 |
Hb_185830_060 |
0.1156358457 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 5 |
Hb_001301_280 |
0.1156553231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000365_010 |
0.1234435779 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 7 |
Hb_020390_010 |
0.1250557049 |
- |
- |
PREDICTED: expansin-A13 [Jatropha curcas] |
| 8 |
Hb_000375_300 |
0.1262397236 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 9 |
Hb_000288_020 |
0.1284489809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002615_030 |
0.1290585104 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 11 |
Hb_000599_210 |
0.1293584062 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 12 |
Hb_002193_060 |
0.1296993949 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 13 |
Hb_005993_010 |
0.1299661162 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 14 |
Hb_004607_070 |
0.1331513653 |
- |
- |
integral membrane protein, putative [Ricinus communis] |
| 15 |
Hb_011344_200 |
0.1354140753 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |
| 16 |
Hb_000049_160 |
0.1357265555 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 17 |
Hb_000012_110 |
0.1383392692 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 18 |
Hb_000116_560 |
0.1391456844 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 19 |
Hb_000438_140 |
0.1391538798 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 20 |
Hb_004158_020 |
0.1393343296 |
- |
- |
Rab1 [Hevea brasiliensis] |