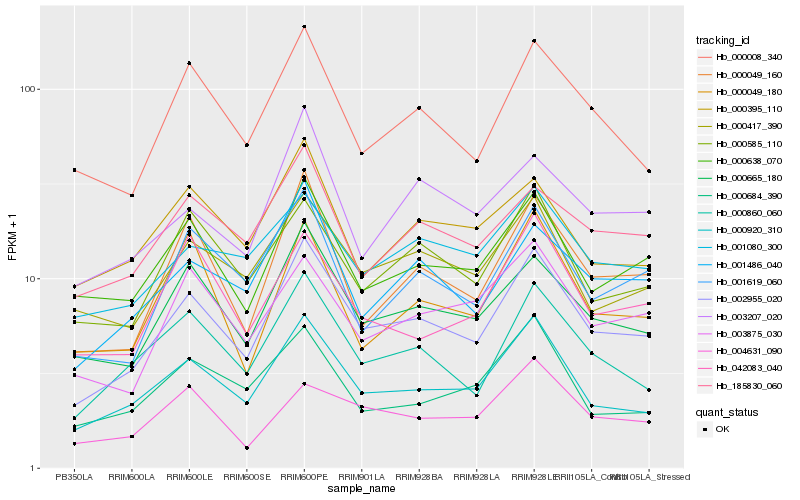
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002955_020 |
0.0 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_000920_310 |
0.0741278731 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 3 |
Hb_000665_180 |
0.0851545822 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 4 |
Hb_000860_060 |
0.0893331042 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 5 |
Hb_000049_160 |
0.0960798215 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 6 |
Hb_001486_040 |
0.098838576 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000638_070 |
0.1000780972 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 8 |
Hb_000008_340 |
0.1003845722 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 9 |
Hb_000417_390 |
0.1018376584 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 10 |
Hb_001619_060 |
0.1059769238 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000585_110 |
0.107541136 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 12 |
Hb_185830_060 |
0.1131702633 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 13 |
Hb_001080_300 |
0.1138222269 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 14 |
Hb_003875_030 |
0.1155330464 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 15 |
Hb_003207_020 |
0.1181122798 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 16 |
Hb_000395_110 |
0.1206101413 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 17 |
Hb_000684_390 |
0.1211135618 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 18 |
Hb_042083_040 |
0.1218973065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004631_090 |
0.1227308388 |
- |
- |
PREDICTED: probable glucan 1,3-beta-glucosidase A [Jatropha curcas] |
| 20 |
Hb_000049_180 |
0.1230129357 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |