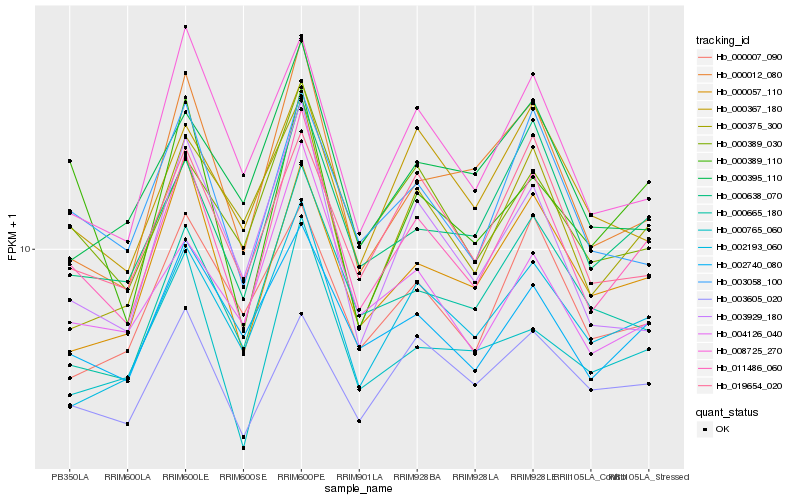
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011486_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000638_070 |
0.0905021516 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 3 |
Hb_003058_100 |
0.0944961595 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_002740_080 |
0.0952518007 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 5 |
Hb_000012_080 |
0.0963993986 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 6 |
Hb_000057_110 |
0.0973612423 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 7 |
Hb_000389_030 |
0.0974050695 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 8 |
Hb_002193_060 |
0.0977434394 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 9 |
Hb_008725_270 |
0.10334996 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 10 |
Hb_019654_020 |
0.1035720494 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 11 |
Hb_000665_180 |
0.1064583458 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_004126_040 |
0.1089372802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000765_060 |
0.1110208881 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 14 |
Hb_000375_300 |
0.1116681786 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 15 |
Hb_003929_180 |
0.1122446753 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 16 |
Hb_000367_180 |
0.112562313 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 17 |
Hb_000007_090 |
0.114598105 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 18 |
Hb_000395_110 |
0.1167537032 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 19 |
Hb_003605_020 |
0.1167983028 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 20 |
Hb_000389_110 |
0.118047045 |
- |
- |
adenosine kinase, putative [Ricinus communis] |