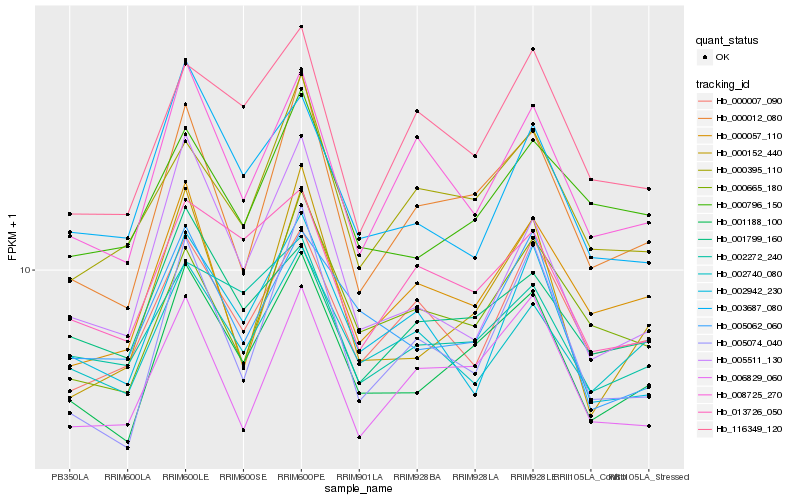
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001188_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 2 |
Hb_000012_080 |
0.0884295316 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 3 |
Hb_006829_060 |
0.1104214672 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000152_440 |
0.1107796453 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 5 |
Hb_005062_060 |
0.1119738958 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 6 |
Hb_002272_240 |
0.1147528437 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_000796_150 |
0.1154839013 |
- |
- |
PREDICTED: uncharacterized protein LOC105136996 isoform X1 [Populus euphratica] |
| 8 |
Hb_008725_270 |
0.1163306442 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 9 |
Hb_001799_160 |
0.1164404128 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_000665_180 |
0.1183931683 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 11 |
Hb_000395_110 |
0.1187570827 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 12 |
Hb_005074_040 |
0.1203654739 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_013726_050 |
0.1204744227 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 14 |
Hb_000057_110 |
0.1211106227 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 15 |
Hb_003687_080 |
0.1213082613 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 16 |
Hb_002740_080 |
0.1220430327 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 17 |
Hb_005511_130 |
0.1244724856 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 18 |
Hb_000007_090 |
0.1252494296 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 19 |
Hb_116349_120 |
0.1254173705 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_002942_230 |
0.1261268393 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |