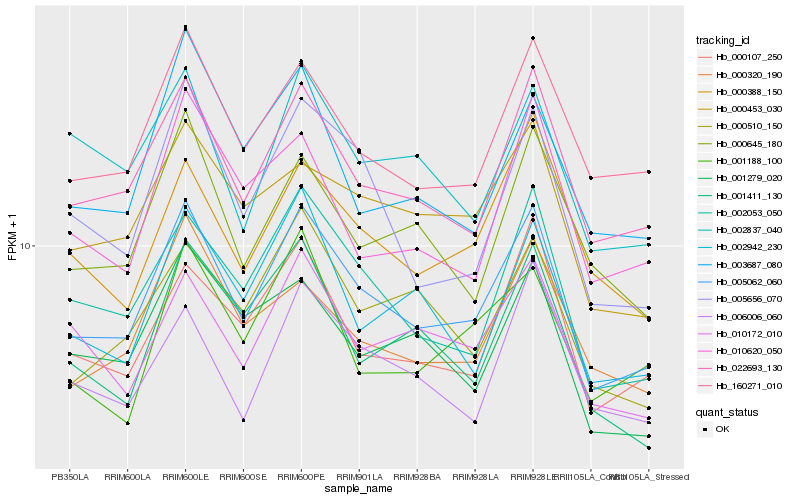
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005062_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 2 |
Hb_022693_130 |
0.0673585456 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_002053_050 |
0.0798993598 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 4 |
Hb_160271_010 |
0.0883905156 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_005656_070 |
0.0914215533 |
- |
- |
PREDICTED: cryptochrome DASH, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_000107_250 |
0.0927318997 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 7 |
Hb_000453_030 |
0.0949542244 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 8 |
Hb_000388_150 |
0.0992435319 |
- |
- |
hypothetical protein JCGZ_21651 [Jatropha curcas] |
| 9 |
Hb_010172_010 |
0.1009311415 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 10 |
Hb_000645_180 |
0.1018636487 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 11 |
Hb_002942_230 |
0.1022100051 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 12 |
Hb_000320_190 |
0.1036089398 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_006006_060 |
0.1047452197 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000510_150 |
0.1050653288 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001279_020 |
0.1098772242 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 16 |
Hb_010620_050 |
0.1103924005 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 17 |
Hb_001411_130 |
0.1111708008 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001188_100 |
0.1119738958 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 19 |
Hb_003687_080 |
0.1120388691 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 20 |
Hb_002837_040 |
0.1128541117 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |