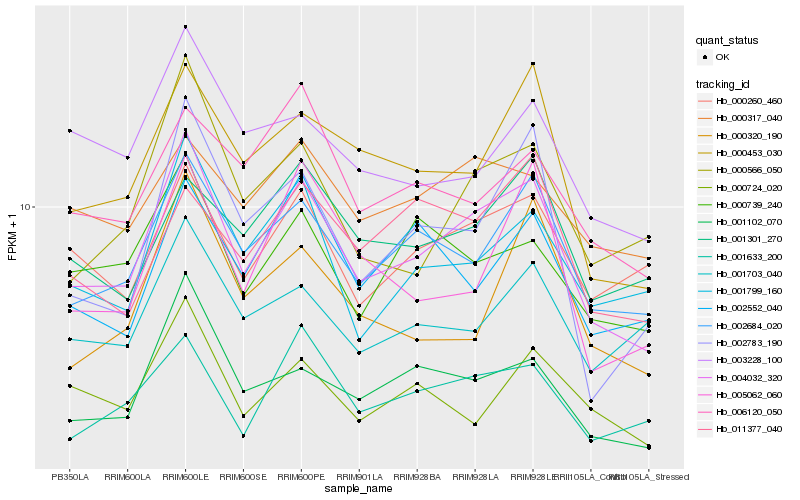
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635153 [Jatropha curcas] |
| 2 |
Hb_002684_020 |
0.0857186722 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 3 |
Hb_001102_070 |
0.0944897155 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000453_030 |
0.0953744229 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 5 |
Hb_001799_160 |
0.100698564 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_001633_200 |
0.1044187492 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 7 |
Hb_000566_050 |
0.1050530787 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 8 |
Hb_000260_460 |
0.1058448388 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000739_240 |
0.1122194756 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002783_190 |
0.1132375593 |
- |
- |
PREDICTED: uncharacterized protein LOC105635339 [Jatropha curcas] |
| 11 |
Hb_001301_270 |
0.1138844736 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 12 |
Hb_005062_060 |
0.1145769272 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 13 |
Hb_000317_040 |
0.1154878015 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 14 |
Hb_006120_050 |
0.1160190824 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 15 |
Hb_000724_020 |
0.1170238345 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 16 |
Hb_001703_040 |
0.1172970557 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 17 |
Hb_011377_040 |
0.1177781849 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 18 |
Hb_003228_100 |
0.1182418421 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 19 |
Hb_000320_190 |
0.1183000143 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002552_040 |
0.118434762 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |