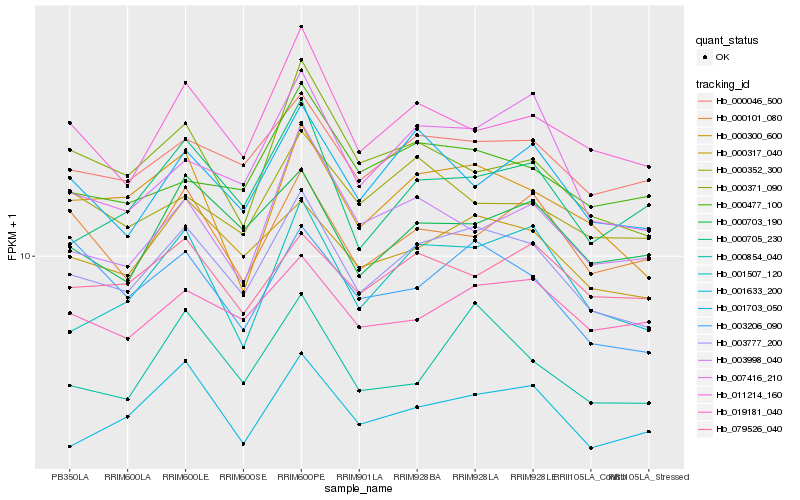
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003777_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 2 |
Hb_000317_040 |
0.063917127 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_000046_500 |
0.0738600854 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 4 |
Hb_000300_600 |
0.0749098331 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 5 |
Hb_011214_160 |
0.0842901859 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 6 |
Hb_001703_050 |
0.0846437025 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 7 |
Hb_001507_120 |
0.085437425 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_000703_190 |
0.0879043555 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_019181_040 |
0.089136838 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 10 |
Hb_000705_230 |
0.0905577734 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 11 |
Hb_000854_040 |
0.092556329 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 12 |
Hb_007416_210 |
0.0925922615 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 13 |
Hb_001633_200 |
0.0928158341 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 14 |
Hb_000101_080 |
0.0940411024 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 15 |
Hb_079526_040 |
0.0941961838 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 16 |
Hb_000477_100 |
0.094440819 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 17 |
Hb_000371_090 |
0.0956659475 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 18 |
Hb_003206_090 |
0.0958970145 |
- |
- |
PREDICTED: protein trichome birefringence-like 5 [Jatropha curcas] |
| 19 |
Hb_000352_300 |
0.0975101973 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 20 |
Hb_003998_040 |
0.0979775138 |
- |
- |
organic anion transporter, putative [Ricinus communis] |