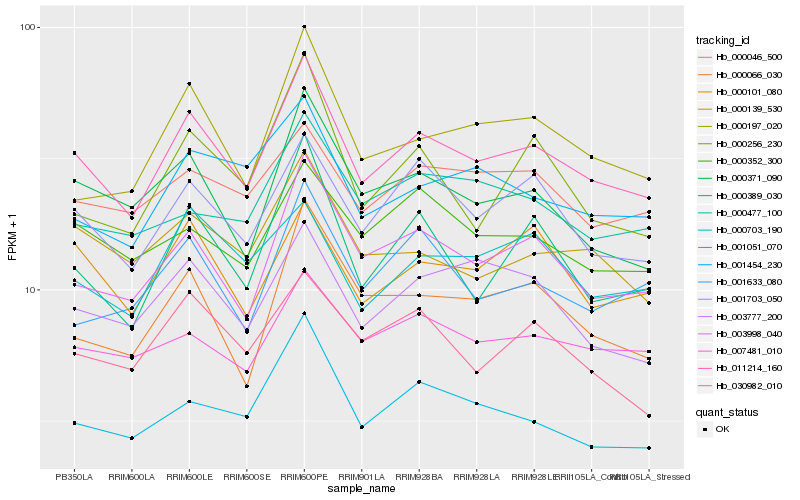
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011214_160 |
0.0 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 2 |
Hb_003998_040 |
0.0652029736 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 3 |
Hb_001703_050 |
0.0699721213 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 4 |
Hb_000371_090 |
0.0771254679 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 5 |
Hb_001051_070 |
0.079644704 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000197_020 |
0.079861614 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 7 |
Hb_000139_530 |
0.0800232414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000389_030 |
0.0814375977 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 9 |
Hb_000101_080 |
0.0830663483 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 10 |
Hb_007481_010 |
0.0842499518 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 11 |
Hb_003777_200 |
0.0842901859 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 12 |
Hb_000352_300 |
0.085354882 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 13 |
Hb_001454_230 |
0.0857884096 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 14 |
Hb_030982_010 |
0.0861824214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000066_030 |
0.0864571064 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000703_190 |
0.0869738402 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000046_500 |
0.0870015039 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 18 |
Hb_001633_080 |
0.0903394552 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 19 |
Hb_000256_230 |
0.0914326506 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 20 |
Hb_000477_100 |
0.0918242318 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |