| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
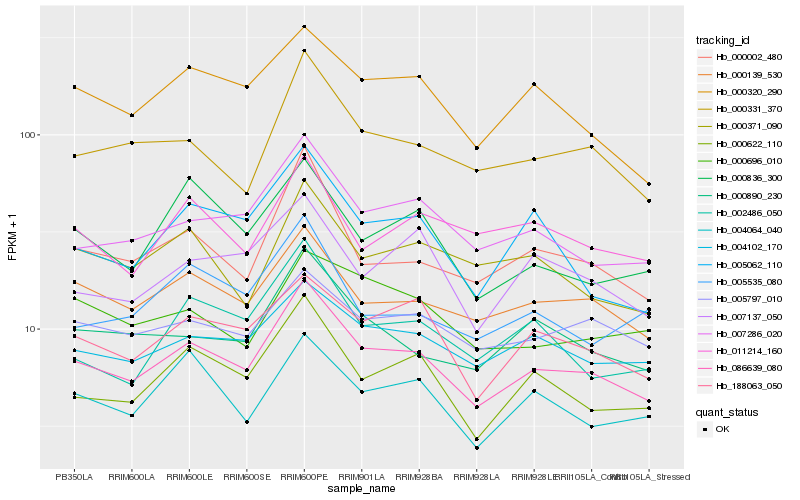
Hb_086639_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000139_530 |
0.0744364091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007286_020 |
0.0786722709 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 4 |
Hb_000622_110 |
0.0787096503 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 5 |
Hb_000002_480 |
0.0801494652 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000331_370 |
0.0808872999 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 7 |
Hb_004102_170 |
0.0835441375 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 8 |
Hb_000890_230 |
0.0843665179 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 9 |
Hb_000320_290 |
0.0860028561 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 10 |
Hb_188063_050 |
0.0868343826 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 11 |
Hb_005797_010 |
0.0942183445 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 12 |
Hb_004064_040 |
0.0974818551 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 13 |
Hb_005062_110 |
0.0991165826 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 14 |
Hb_000371_090 |
0.099194258 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 15 |
Hb_002486_050 |
0.0999471519 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_007137_050 |
0.1008006811 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_011214_160 |
0.1032401359 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 18 |
Hb_000836_300 |
0.1041668681 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 19 |
Hb_000696_010 |
0.104390929 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 20 |
Hb_005535_080 |
0.1044574499 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |