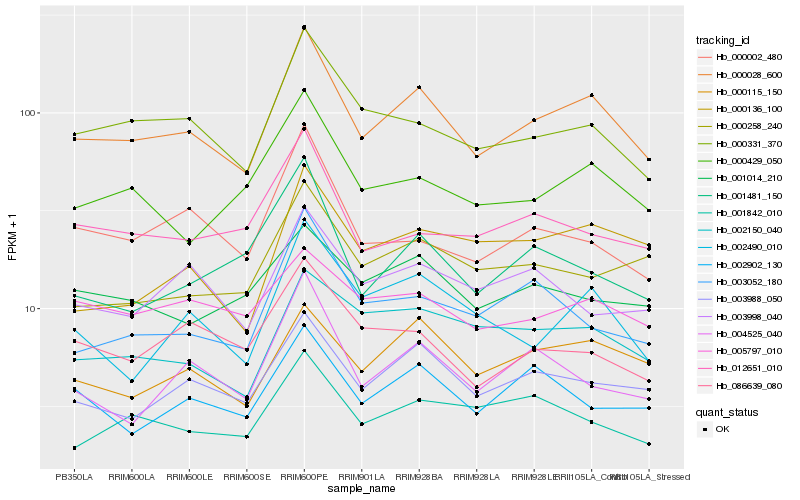
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_600 |
0.0 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 2 |
Hb_003988_050 |
0.0902224374 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 3 |
Hb_000331_370 |
0.0968254899 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 4 |
Hb_000115_150 |
0.0988032043 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 5 |
Hb_000429_050 |
0.106491415 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002490_010 |
0.112004263 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |
| 7 |
Hb_002150_040 |
0.114571782 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 8 |
Hb_002902_130 |
0.1154025861 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 9 |
Hb_000258_240 |
0.1157136152 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 10 |
Hb_000002_480 |
0.1163719037 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_012651_010 |
0.1167232714 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 12 |
Hb_004525_040 |
0.1169624371 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 13 |
Hb_001842_010 |
0.1173238988 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_086639_080 |
0.1177054894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003052_180 |
0.1182063943 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 16 |
Hb_003998_040 |
0.1187019981 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 17 |
Hb_005797_010 |
0.1191188072 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 18 |
Hb_001481_150 |
0.1194630464 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 19 |
Hb_001014_210 |
0.1204834269 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 20 |
Hb_000136_100 |
0.1206644162 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |