| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
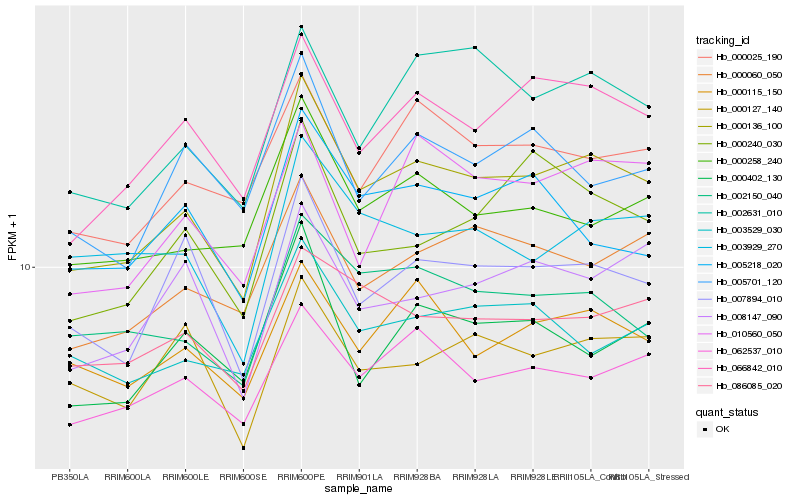
Hb_000136_100 |
0.0 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 2 |
Hb_066842_010 |
0.0870685721 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007894_010 |
0.0964012543 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 4 |
Hb_000060_050 |
0.0964588098 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 5 |
Hb_000258_240 |
0.0978491318 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 6 |
Hb_062537_010 |
0.0994160612 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 7 |
Hb_002150_040 |
0.0998329502 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000115_150 |
0.1016173571 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 9 |
Hb_000025_190 |
0.1017850824 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 10 |
Hb_086085_020 |
0.1027085534 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 11 |
Hb_002631_010 |
0.1035388907 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 12 |
Hb_003929_270 |
0.1063531374 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_003529_030 |
0.1074922405 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 14 |
Hb_000402_130 |
0.107672142 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 15 |
Hb_000240_030 |
0.1103263907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000127_140 |
0.1104828193 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 17 |
Hb_008147_090 |
0.1106602191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005218_020 |
0.1115845156 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 19 |
Hb_005701_120 |
0.1127270179 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 20 |
Hb_010560_050 |
0.1134539601 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |