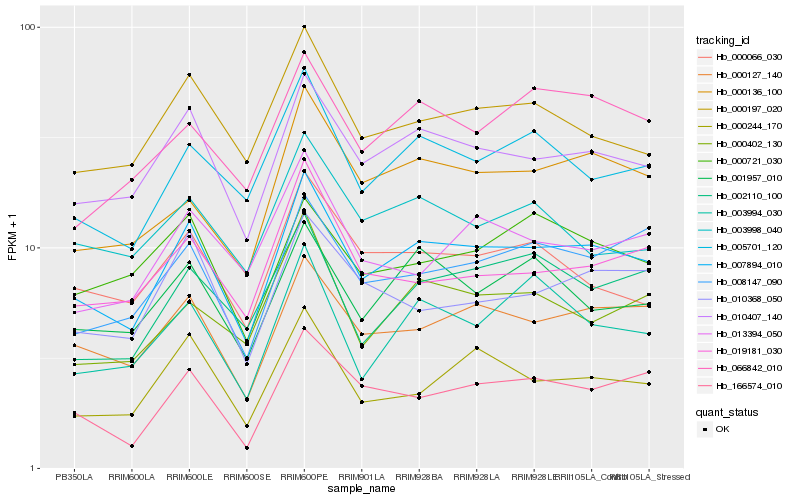
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 2 |
Hb_010407_140 |
0.0594120741 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 3 |
Hb_000127_140 |
0.07027183 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 4 |
Hb_005701_120 |
0.0869355048 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 5 |
Hb_000197_020 |
0.089223174 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 6 |
Hb_000136_100 |
0.0964012543 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 7 |
Hb_001957_010 |
0.0964103195 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000244_170 |
0.0971995184 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 9 |
Hb_166574_010 |
0.0972480677 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 10 |
Hb_008147_090 |
0.0974400792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000066_030 |
0.098658613 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 12 |
Hb_010368_050 |
0.1020860675 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 13 |
Hb_000402_130 |
0.103740316 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 14 |
Hb_019181_030 |
0.1042915222 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 15 |
Hb_002110_100 |
0.1046807339 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003994_030 |
0.1062526689 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 17 |
Hb_003998_040 |
0.1063311672 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 18 |
Hb_000721_030 |
0.1083344108 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 19 |
Hb_066842_010 |
0.1084364756 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_013394_050 |
0.1085622671 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |