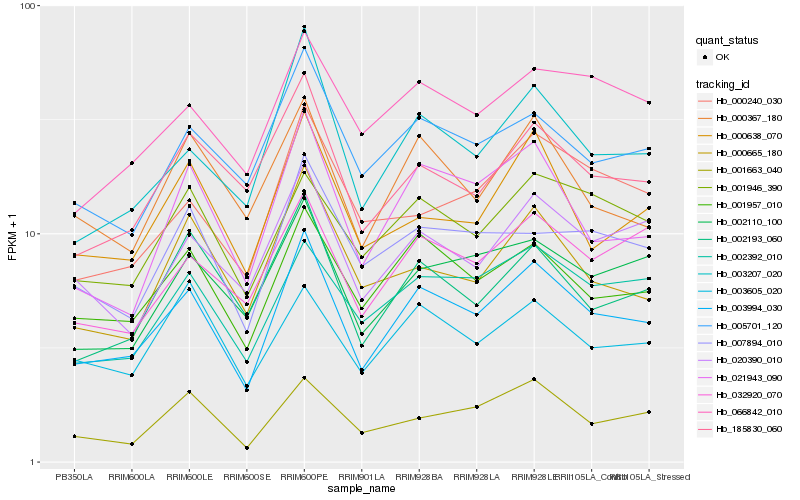
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003994_030 |
0.0 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 2 |
Hb_001957_010 |
0.0816281611 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003207_020 |
0.0857079854 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 4 |
Hb_021943_090 |
0.0898244623 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 5 |
Hb_002110_100 |
0.1008553291 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002193_060 |
0.100857803 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 7 |
Hb_005701_120 |
0.1010786573 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 8 |
Hb_002392_010 |
0.1012288189 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 9 |
Hb_020390_010 |
0.1016781036 |
- |
- |
PREDICTED: expansin-A13 [Jatropha curcas] |
| 10 |
Hb_185830_060 |
0.102237098 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 11 |
Hb_000367_180 |
0.1028554844 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 12 |
Hb_000638_070 |
0.1035910338 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 13 |
Hb_001663_040 |
0.1037050573 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 14 |
Hb_032920_070 |
0.105819746 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 15 |
Hb_007894_010 |
0.1062526689 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 16 |
Hb_001946_390 |
0.106782871 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 17 |
Hb_066842_010 |
0.1074585091 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000665_180 |
0.1091857369 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_003605_020 |
0.1096023864 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 20 |
Hb_000240_030 |
0.1107202587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |