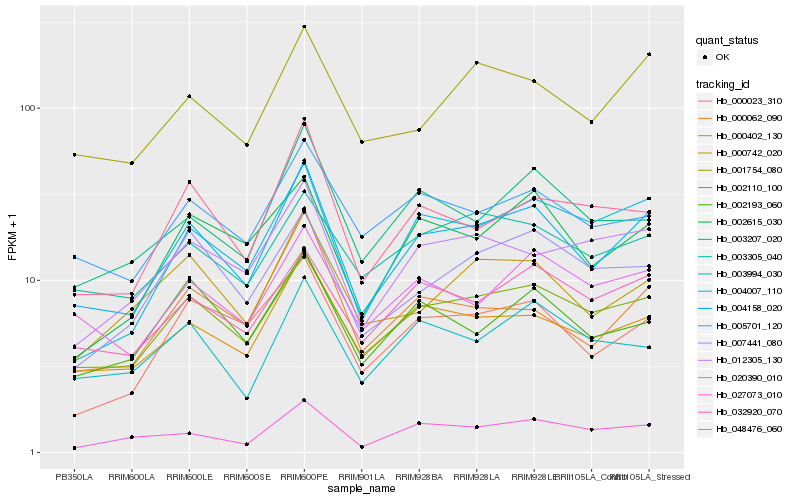
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004158_020 |
0.0 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 2 |
Hb_002110_100 |
0.0772422891 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000402_130 |
0.0937973488 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 4 |
Hb_032920_070 |
0.1033499999 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 5 |
Hb_001754_080 |
0.1055542785 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 6 |
Hb_004007_110 |
0.1059746433 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 7 |
Hb_000023_310 |
0.1075483636 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 8 |
Hb_020390_010 |
0.1109666244 |
- |
- |
PREDICTED: expansin-A13 [Jatropha curcas] |
| 9 |
Hb_005701_120 |
0.1111569652 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 10 |
Hb_002615_030 |
0.1123715962 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 11 |
Hb_003207_020 |
0.1133924913 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 12 |
Hb_000742_020 |
0.1142572713 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 13 |
Hb_048476_060 |
0.1159582296 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_002193_060 |
0.1161479642 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 15 |
Hb_003305_040 |
0.1176533231 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 16 |
Hb_012305_130 |
0.1177569976 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 17 |
Hb_027073_010 |
0.1191817336 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 18 |
Hb_000062_090 |
0.1192241221 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003994_030 |
0.1197185358 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 20 |
Hb_007441_080 |
0.1205916746 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |