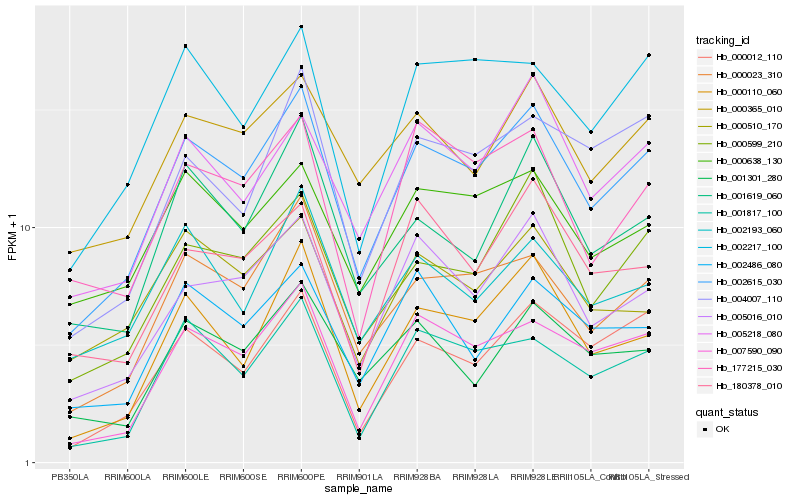
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002615_030 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 2 |
Hb_000023_310 |
0.0754788555 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 3 |
Hb_005016_010 |
0.0844538676 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 4 |
Hb_007590_090 |
0.0852576245 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 5 |
Hb_000599_210 |
0.0871883376 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000110_060 |
0.0922371514 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 7 |
Hb_000365_010 |
0.0935565661 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 8 |
Hb_005218_080 |
0.0949536552 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000012_110 |
0.0975577884 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 10 |
Hb_001817_100 |
0.0979104804 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 11 |
Hb_002217_100 |
0.0996307308 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_001619_060 |
0.1028736003 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000638_130 |
0.1031748296 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 14 |
Hb_001301_280 |
0.1055704682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004007_110 |
0.1084127198 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 16 |
Hb_002486_080 |
0.1086772623 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 17 |
Hb_002193_060 |
0.1092813011 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 18 |
Hb_000510_170 |
0.1099431469 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 19 |
Hb_177215_030 |
0.1101150417 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_180378_010 |
0.1108908531 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |