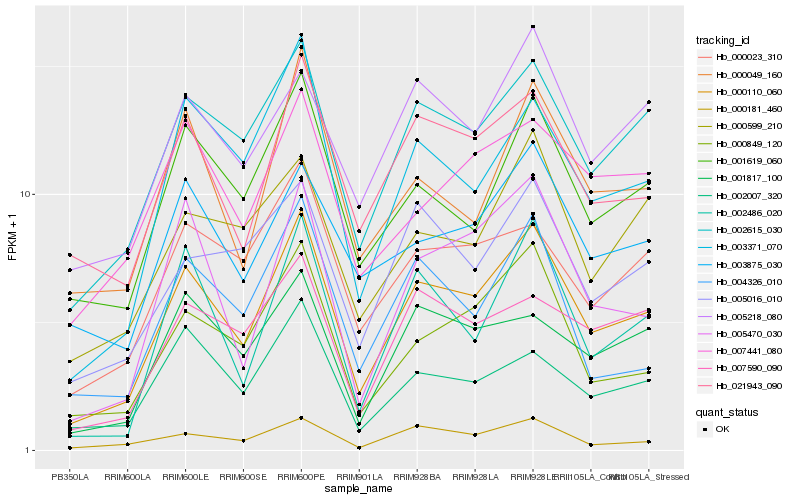
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 2 |
Hb_002615_030 |
0.0922371514 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 3 |
Hb_003371_070 |
0.1026911135 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000849_120 |
0.1104048081 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 5 |
Hb_001619_060 |
0.1109682679 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000181_460 |
0.1119362402 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 7 |
Hb_021943_090 |
0.1127706806 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 8 |
Hb_005470_030 |
0.1150729221 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002007_320 |
0.1181205952 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 10 |
Hb_000049_160 |
0.1185905548 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 11 |
Hb_002486_020 |
0.1190258016 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000023_310 |
0.1195330346 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 13 |
Hb_001817_100 |
0.120015228 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 14 |
Hb_007590_090 |
0.1240762233 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 15 |
Hb_005016_010 |
0.1268531704 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 16 |
Hb_003875_030 |
0.1279979498 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 17 |
Hb_005218_080 |
0.1285649909 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000599_210 |
0.1310193553 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 19 |
Hb_007441_080 |
0.1320927045 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 20 |
Hb_004326_010 |
0.1323029337 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |