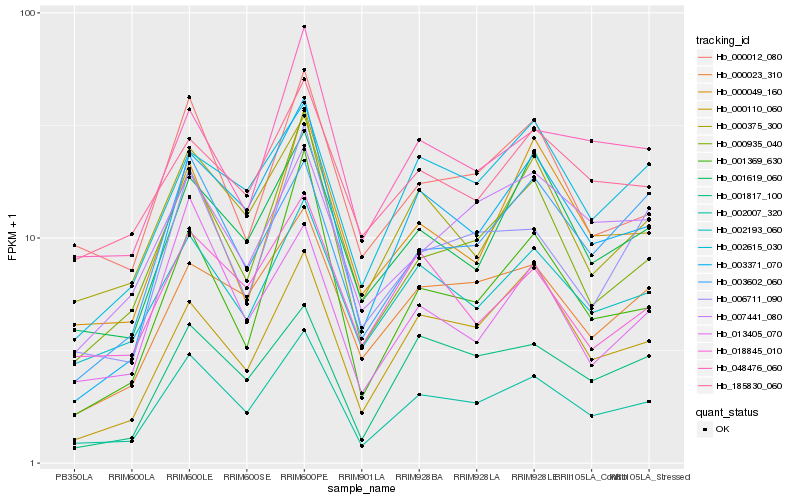
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002007_320 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 2 |
Hb_002193_060 |
0.0891677644 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 3 |
Hb_003371_070 |
0.0902307105 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000375_300 |
0.0930872045 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 5 |
Hb_000935_040 |
0.09323892 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 6 |
Hb_000012_080 |
0.0966871887 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 7 |
Hb_000023_310 |
0.1039062789 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 8 |
Hb_006711_090 |
0.1054811283 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 9 |
Hb_001619_060 |
0.1063472296 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001817_100 |
0.1076649522 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 11 |
Hb_048476_060 |
0.1110720227 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_003602_060 |
0.1135730245 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 13 |
Hb_018845_010 |
0.116637531 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 14 |
Hb_001369_630 |
0.1167229623 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 15 |
Hb_000110_060 |
0.1181205952 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 16 |
Hb_002615_030 |
0.1184626795 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 17 |
Hb_185830_060 |
0.120941967 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 18 |
Hb_007441_080 |
0.1218033738 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 19 |
Hb_013405_070 |
0.1219271816 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 20 |
Hb_000049_160 |
0.1236860753 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |